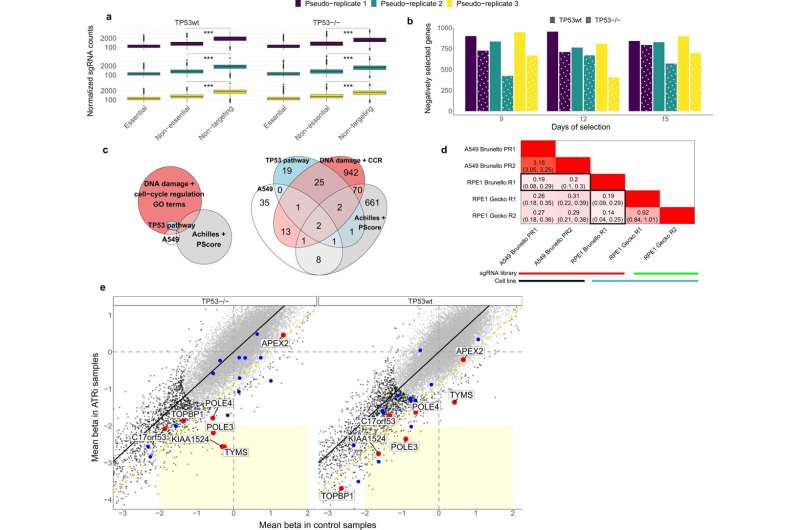
A TP53 wild-type background can confound estimates of gene selection in genetic screens. a Boxplots showing the pooled normalized sgRNA counts per sample (essential and non-essential genes, and non-targeting sgRNAs; 15 day samples are shown). Tested using 1-tailed Mann-Whitney. *** denotes a p <2.2e-16. No adjustments were made for multiple comparisons. n = 7300 independent sgRNAs examined over six independent experiments. b Barplot showing the number of genes that are negatively (beta score<0) selected, per sample used in this study. Beta score significance: FDR < 0.25. c Venn (left) and corresponding Euler (right) diagrams of the overlap of genes between four sets: genes negatively selected exclusively in TP53wt in our samples (A549), genes negatively selected exclusively in TP53wt in Project Achilles and Score (Achilles + Score), top-50 TP53-interactors (TP53 pathway), and genes included in 19 GO terms related to DNA damage and cell-cycle regulation that we found enriched with genes from the A549 set. d Results of the analysis of overlap between different cell lines and/or sgRNA libraries detailed in Supplementary Text 1a: heatmap shows the log2 odds ratio of the overlap of genes negatively selected exclusively in TP53wt, between different experiments. R: Replicate, PR: Pseudo-replicate. Darker shades of red indicate higher overlap. Black rectangles highlight the overlap between RPE1 Brunello dataset with others. e Comparison between TP53-isogenic cell lines to assess biases in identifying conditional essentiality from genetic screens. x and y axes represent the standardized beta scores (Z-scores) for genes either in the control samples (incl. doxycycline-treated; pseudo-replicates 1 and 2), or in the doxycycline+ATRi treated samples, respectively, averaged across later time points and pseudo-replicates. Coordinate axes were capped in order to zoom on the region of interest. The EM clustering identified two gene clusters as the most likely model, represented by black and gray dots. Black line represents the best fit linear model. The yellow dashed diagonal line represents −2 standard deviations (SD) of the Z-score difference. The light yellow rectangle delimits the tentative significance area containing genes negatively selected in the treatment, but not selected in the control sample (i.e., potentially synthetic lethal with ATRi). The top-20 validated ATRi-sensitizing genes are highlighted with color, and the top-7 (red) are further labelled. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-32285-1
CRISPR/Cas9 is a precise gene editing technique whose development by Jennifer A. Doudna and Emmanuelle Charpentier was recognized with the 2020 Nobel Prize in Chemistry. Commonly known as "genetic scissors," CRISPR allows the introduction of the desired DNA sequence into (virtually) any spot of the genome, thus modifying or inactivating a gene. This technique is widely used in biomedical research and some CRISPR-based therapies are in clinical trials for the treatment of human blood disorders, some types of cancer and HIV, among other conditions.
Scientists at IRB Barcelona, led by ICREA researcher Dr. Fran Supek, have now reported that, depending on the targeted spot of the human genome, CRISPR gene editing can give rise to cell toxicity and genomic instability. This unwanted effect is mediated by the linchpin tumor suppressor protein p53 and is determined by the DNA sequence near the editing point and various epigenetic factors in the surrounding region.
Using computational methods, researchers in the Genome Data Science lab have analyzed the most popular CRISPR library designed for human cells and have detected 3,300 targeted spots that show strong toxic effects. The work, published in Nature Communications, also reports that around 15% of the human genes contain at least one toxic editing point.
"Our work addresses an important issue with TP53-associated toxicity of Cas9, which was a matter of some controversy recently, and it also provides guidelines on how to sidestep the problem. Avoiding editing in these 'risky' spots would not only make CRISPR editing more efficient but, more importantly, safer," Dr. Supek explains.
A specific gene can be edited in a variety of positions. "The regions of the gene that are important for regulation or have certain epigenetic markers are the ones most likely to trigger the p53 response and should, therefore, be avoided as a general recommendation," says Dr. Miguel-Martin Álvarez, a lead researcher on the study.
p53-mediated toxicity and tumorigenesis
p53 is a protein known as the guardian of the genome. It detects DNA damage and leads the cells to stop dividing and can cause programmed death, thus preventing them from reproducing and expanding the "mistakes" in their DNA. Therefore, p53 underlies a natural protection mechanism against cancer and other DNA damage-related complications.
CRISPR gene editing often requires cutting both DNA strands. In some cases, this manipulation can trigger a p53 response, in which edited cells can be "tagged" as damaged and are then eliminated, thus reducing the efficiency of the gene editing process.
However, the main complication regarding p53 and gene editing is that cells that overcome CRISPR editing might do so precisely because of defective p53 functioning. That is to say, these cells may be less able to detect DNA damage and/or tag cells for programmed death. As a result, the gene editing procedure could end up favoring cell populations that have unstable genomes, meaning they are prone to accumulating further mutations, thus increasing the risk of developing malignancies.
"This unwanted consequence might incur a risk of genomic instability, which is highly undesirable in the context of ex vivo CRISPR therapies, in which cells from a patient are edited in the lab and reintroduced back into the patient. We hope that our study provides some guidelines on how to design safer CRISPR reagents, and encourages further research into this issue," concludes Dr. Supek.
More information: Miguel M. Álvarez et al, TP53-dependent toxicity of CRISPR/Cas9 cuts is differential across genomic loci and can confound genetic screening, Nature Communications (2022). DOI: 10.1038/s41467-022-32285-1
Journal information: Nature Communications