Credit: Pixabay/CC0 Public Domain
Many people suffer from eye diseases that can lead to blindness in the worst cases. Eye-related diseases including cataracts, glaucoma and macular degeneration are well described; nevertheless, the underlying causative genes are frequently unknown. A team of scientists from Frankfurt and Dresden has now set out to identify some of these undiscovered genes in mammals that take over functions in the eye. They discovered 15 previously unknown eye-related genes in their large-scale genomic analysis and confirmed 14 more genes with known roles in the eye. The study is ground-breaking as it uses genome analysis to predict gene function. It lays the foundation for further research related to vision and the eye.
Reduced vision greatly affects people's everyday life and life quality. In contrast, some animals do not rely on eyesight as their primary sense, and so vision was diminished during the course of evolution. Most prominent examples of mammals with poor eyesight are the echolocating big brown bat, and, among rodents, blind moles and the naked mole rat. On the other hand, closely related species can see well: non-echolocating flying foxes and rodents including rats and guinea pigs.
A new study, published in the journal eLife, compared 49 genomes of mammals with good and poor eyesight. The study is a collaboration between the LOEWE Center for Translational Biodiversity Genomics and the Senckenberg Nature Research Society in Frankfurt as well as the Max Planck Institute for Molecular Cell Biology and Genetics in Dresden and the Center for Regenerative Therapies of the Technical University Dresden.
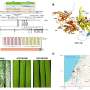
The scientists compared 49 genomes of mammals with good and poor eyesight. "For every species, we investigated genes for their intactness. If a gene is intact, it will probably perform functions. In contrast, broken genes are usually connected to a loss of function. This then results in a so-called 'evolutionary loss pattern,'" explains the first author of the paper, Dr. Henrike Indrischek from the Max Planck Institute for Molecular Cell Biology and Genetics. "It is very informative and interesting to see how the losses of a gene are distributed among different animal species. If a gene is deleted in the blind golden mole, but intact in the elephant, which is its closest relative included in this study, this may indicate a function in the eye. Genes on our list with a potential eye-related function are required to be lost at least three times independently in different animal species with poor eyesight."
The team particularly considered one of the 29 genes likely involved in vision: SERPINE3. Performing experiments in zebrafish, they were able to demonstrate that deletion of this gene can lead to structural changes in the eye and retina. "Zebrafish are used in eye research as they largely rely on vision just as humans and reproduce quickly," explains Indrischek. "Other studies also link SERPINE3 to genetic variants that influence traits in the human eye. Therefore, we suspect that disruption of this gene in humans can contribute to features of the eye and age-related eye diseases."
The publication is one of few studies that use gene-loss patterns to predict gene function. "This enables us to track down previously unknown eye-related genes," says study leader Prof. Dr. Michael Hiller from the LOEWE Center for Translational Biodiversity Genomics. "In doing so, we combine comparative genomics with experiments in model organisms to identify those genes and predict their functions. During our research, we came across other exciting genes besides SERPINE3, that have not been studied so far. They likely contribute to the fact that eyesight differs across animal species. Here lies potential for future research on eye diseases."
More information: Henrike Indrischek et al, Vision-related convergent gene losses reveal SERPINE3's unknown role in the eye, eLife (2022). DOI: 10.7554/eLife.77999
Journal information: eLife
Provided by Senckenberg Forschungsinstitut und Naturmuseen