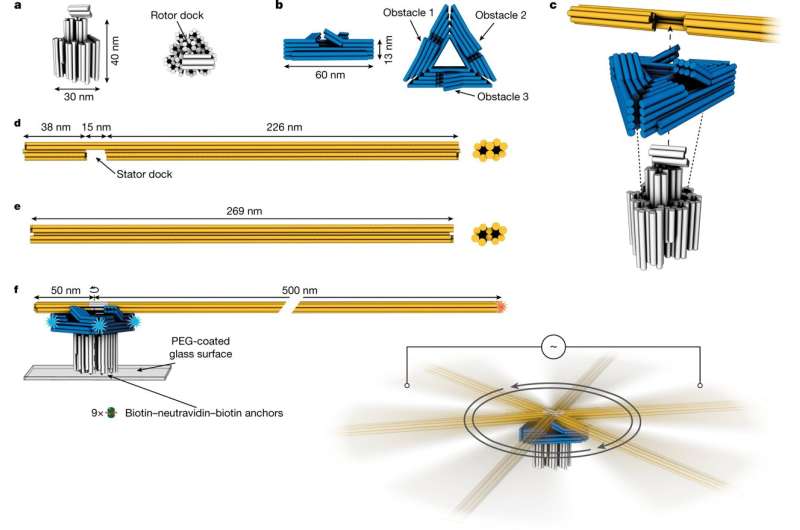
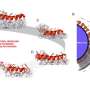
Motor design and experimental setup. a,b, Schematics of a pedestal and a triangular platform, respectively. Cylinders indicate DNA double helices. c, Schematic illustration of motor assembly steps. d,e, Rotor arm components. f, Left, schematic illustration of the experimental setup for observing motor dynamics in an inverted TIRF microscope. The pedestal is fixed through several biotin–neutravidin linkages to a microscope coverslip. Orange star, Cy5 dyes. Blue stars, labeling positions for DNA-PAINT imager strands. Right, two platinum electrodes are immersed in the liquid chamber from above and connected to a function generator generating a square-wave alternating current to create a fixed-axis energetic modulation that acts on all motors. Credit: Nature (2022). DOI: 10.1038/s41586-022-04910-y
A research team led by the Technical University of Munich (TUM) has succeeded for the first time in producing a molecular electric motor using the DNA origami method. The tiny machine made of genetic material self-assembles and converts electrical energy into kinetic energy. The new nanomotors can be switched on and off, and the researchers can control the rotation speed and rotational direction.
Be it in our cars, drills or automatic coffee grinders—motors help us perform work in our everyday lives to accomplish a wide variety of tasks. On a much smaller scale, natural molecular motors perform vital tasks in our bodies. For instance, a motor protein known as ATP synthase produces the molecule adenosine triphosphate (ATP), which our body uses for short-term storage and transfer of energy.
While natural molecular motors are essential, it has been quite difficult to recreate motors on this scale with mechanical properties roughly similar to those of natural molecular motors like ATP synthase. A research team has now constructed a working nanoscale molecular rotary motor using the DNA origami method and published their results in Nature. The team was led by Hendrik Dietz, Professor of Biomolecular Nanotechnology at TUM, Friedrich Simmel, Professor of Physics of Synthetic Biological Systems at TUM, and Ramin Golestanian, director at the Max Planck Institute for Dynamics and Self-Organization.
A self-assembling nanomotor
The novel molecular motor consists of DNA—genetic material. The researchers used the DNA origami method to assemble the motor from DNA molecules. This method was invented by Paul Rothemund in 2006 and was later further developed by the research team at TUM. Several long single strands of DNA serve as a basis to which additional DNA strands attach themselves to as counterparts. The DNA sequences are selected in such a way that the attached strands and folds create the desired structures.
"We've been advancing this method of fabrication for many years and can now develop very precise and complex objects, such as molecular switches or hollow bodies that can trap viruses. If you put the DNA strands with the right sequences in solution, the objects self-assemble," says Dietz.
The new nanomotor made of DNA material consists of three components: base, platform and rotor arm. The base is approximately 40 nanometers high and is fixed to a glass plate in solution via chemical bonds on a glass plate. A rotor arm of up to 500 nanometers in length is mounted on the base so that it can rotate. Another component is crucial for the motor to work as intended: a platform that lies between the base and the rotor arm. This platform contains obstacles that influence the movement of the rotor arm. To pass the obstacles and rotate, the rotor arm must bend upward a little, similar to a ratchet.
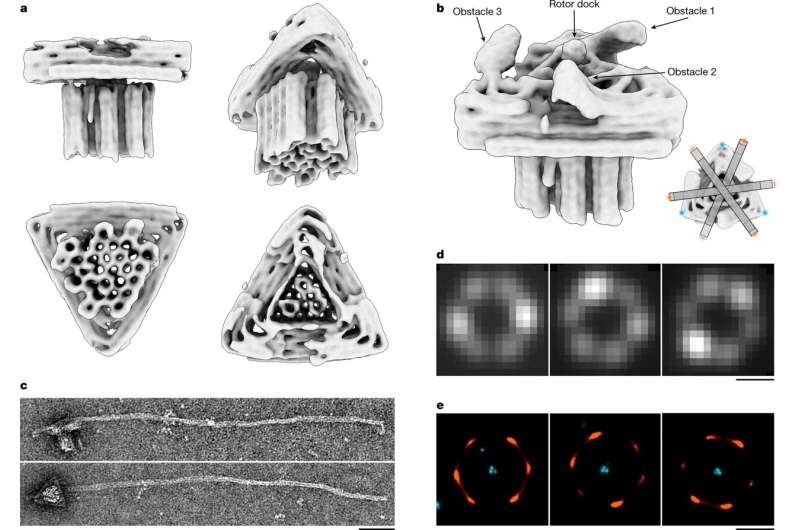
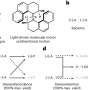
Structural analysis of the DNA origami motor. a, Different views of a 3D electron density map of the motor block determined by means of single-particle cryo-EM (see also Extended Data Fig. 4 and in the Electron Microscopy Data Bank (EMDB) under code EMD-14358). b, Motor block cryo-EM map detail depicted at different density thresholds at which the three obstacles and the rotor dock can be discerned. Inset, schematic showing the six preferred dwelling sites of the rotor arm. c, Exemplary negative-staining TEM images of a motor variant with long rotor arm attached. Scale bar, 50 nm. d, Exemplary single-particle fluorescence images. Scale bar, 500 nm. The images show the standard deviation of the mean intensity per pixel computed over all the frames from recorded TIRF videos. e, DNA-PAINT images showing rotor arm tip positions relative to the triangle platform. Scale bar, 500 nm. Credit: Nature (2022). DOI: 10.1038/s41586-022-04910-y
Targeted movement through AC voltage
Without energy supply, the rotor arms of the motors move randomly in one direction or the other, driven by random collisions with molecules from the surrounding solvent. However, as soon as AC voltage is applied via two electrodes, the rotor arms rotate in a targeted and continuous manner in one direction.
"The new motor has unprecedented mechanical capabilities: It can achieve torques in the range of 10 piconewton times nanometer. And it can generate more energy per second than what's released when two ATP molecules are split," explains Ramin Golestanian, who led the theoretical analysis of the mechanism of the motor.
The targeted movement of the motors results from a superposition of the fluctuating electrical forces with the forces experienced by the rotor arm due to the ratchet obstacles. The underlying mechanism realizes a so-called "flashing Brownian ratchet." The researchers can control the speed and direction of the rotation via the direction of the electric field and also via the frequency and amplitude of the AC voltage.
"The new motor could also have technical applications in the future. If we develop the motor further we could possibly use it in the future to drive user-defined chemical reactions, inspired by how ATP synthase makes ATP driven by rotation. Then, for example, surfaces could be densely coated with such motors. Then you would add starting materials, apply a little AC voltage and the motors produce the desired chemical compound," says Dietz.
More information: Anna-Katharina Pumm et al, A DNA origami rotary ratchet motor, Nature (2022). DOI: 10.1038/s41586-022-04910-y
Journal information: Nature
Provided by Technical University Munich