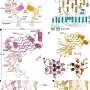
Graphical abstract. Credit: Cell Systems (2022). DOI: 10.1016/j.cels.2022.05.004
Your cells need to get around. For example, immune cells must roam around your body to locate sites of infection, and neurons must migrate to specific positions in the brain during development. But cells do not have eyes to see where they are going. Instead, like a dog sniffing out the source of some delicious smells, a cell figures out how to get to some target by detecting chemicals in its environment through receptors peppered across the cell's surface. For example, the site of an infection will emit certain molecules, and a white blood cell will follow this trail of signals to find their source.
Understanding how cells migrate by reading signals in their environment is a fundamental part of knowing how living systems, from immune cells in the human body to single-celled organisms living in soils, function. New work from the laboratory of Caltech's Matt Thomson, assistant professor of computational biology and Heritage Medical Research Institute Investigator, provides new insights into how cells migrate and respond to information in their environments. The research is described in a paper that appears in the journal Cell Systems on June 8.
Biologists have traditionally understood the process of cell migration with a simple model. In this model, a cell's environment is depicted as a gradient of signal concentrations, with a very high concentration emanating from a source (like the previously mentioned example of infection) that smoothly decreases farther away from the source. As an example, imagine releasing a drop of colored dye into water. The water in the immediate vicinity of where the dye is placed would become brightly colored; with distance from that source, color would gradually decrease in intensity.
But this simple model does not actually replicate what the messy, complex environment inside living tissues looks like.
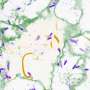
A cell (white blob) successfully migrates to a signal source at the left boundary by following local gradients. However, this simple model does not actually capture what real cellular environments look like, which do not have smooth signaling gradients. Credit: Z. Wang
"If you wanted to engineer cells to do some task in the body for biomedical applications—like killing tumors—that cell is going to have to know how to deal with real environments, not just the simplistic environment of a lab dish," says graduate student Zitong Jerry Wang, the study's first author.
In tissues, cells move around through a tangled web of proteins called the extracellular matrix (ECM). Here, chemical signals do not just float freely—they stick to the ECM itself, creating a signaling environment that does not look like a smooth gradient, but rather a patchy, network-like mess of clustered molecules.
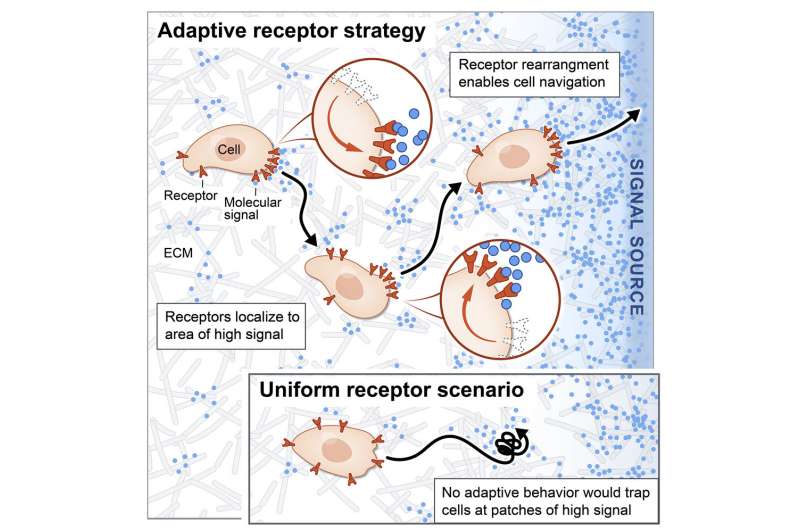
How do cells locate the source of signaling molecules to navigate in the real, messy environment within tissues? The traditional gradient model of cell migration, in which the cell smoothly follows its local signaling concentration gradient, does not work in this realistic environment, because though the cell may discover a patch of relatively high signal concentration, it cannot move away from that local maximum to find the actual source of signals. In other words, the cell gets stuck at local patches of high concentrations, but cannot actually get to where it needs to go. For example, imagine you are attempting to summit a mountain by only moving uphill—you might get stuck at the top of a smaller intermediate hill, because in a real mountainous environment you may need to descend in certain areas in order to reach the highest peak.
To understand how cells deal with this, the team was motivated by experimental observations made in yeast cells showing that when the cells sense pheromones, they rearrange the receptors on their surfaces so that more receptors are placed near areas with a high signal concentration. The team was also intrigued by the fact that dynamic receptor rearrangement had been observed in a variety of systems—certain human cell types like T cells and neurons can rearrange their receptors, and even locusts actively sweep their antennae (containing odor receptors) through space as they move, which significantly improves their ability to navigate to the source of patchy odor plumes.
A cell, tracking local gradients, fails to migrate to the signal source (left boundary) in a tissue environment. Credit: Z. Wang
With this in mind, the team developed a computer model in which cellular receptors could actively redistribute themselves in response to signals, based on known molecular mechanisms for receptor redistribution. In this dynamic model, cells do not get stuck at areas of local concentration, and are able to find the true signal source. Following this receptor optimization, cellular navigation was 30 times more efficient, and the model accurately matched actual cellular behavior observed in tissue. Though receptor rearrangement had been observed in myriad systems, this work is the first to show that it plays a crucial, functional role in cell navigation.
"In an upcoming paper, we describe how the receptor redistribution mechanism we modeled precisely implements what is known as a Bayesian filter, which is a well-known target tracking algorithm that is actively used in robotics today," explains Wang. "So cells in our body could actually be using a similar algorithm for navigation as autonomous vehicles like self-driving cars."
The new model is critical for understanding real cellular systems relevant to human health. "For a long time, people couldn't actually image within tissues, so it was unknown what the tissue environment even looked like," Wang says. "Researchers would take cells out of the body and study how they move in a lab dish, with smoothly diffusing gradients of signals released from a pipette. But now we know this really isn't what is happening in the real environment, which is patchy and complicated. This work has inspired us to actually set up a collaboration with physicians to image more tissue samples to better understand the in vivo environment."
Notably, this research was inspired by principles of neuroscience and how neurons process information about signals in their environments.
With the new model developed in this research, a cell dynamically rearranging its surface receptors is able to navigate local maxima of signal concentrations and locate the source of signals (left boundary). Credit: Z. Wang
"The sensory information an organism receives in its natural environment is highly structured spatiotemporally, meaning it varies over time and in space due to statistical regularities inherent to natural stimuli," Wang says. "Neuroscientists have found that neural sensory processing systems, such as retinal processing and auditory processing, have been adapted to the statistical property of the signals which they are exposed to—the visual or auditory signal in the animal's natural environment."
"We know that a cell also lives in spatially structured environment, so we first constructed statistical models of natural cell environments in both soil and tissue from both imaging data and simulation, then used information theory to ask how a cell's sensory processing system—in this case, distribution of receptors—is related to the statistical structure of the cell's environment.
"We were surprised to find that this general principle from neuroscience applies at the scale of individual cells as well, specifically receptor distributions found on cells drastically improves information acquisition in natural environments. Furthermore, we show the same connection extents to cell navigation. Adaptive rearrangement of receptors observed on cells significantly improves cell navigation but only in natural environments like tissue. This begs the question if there are other aspects of cell biology that can also be better understood when put in the context of a cell's natural habitat, for example strategies of cell-cell communication."
More information: Zitong Jerry Wang, Matt Thomson, Localization of signaling receptors maximizes cellular information acquisition in spatially structured natural environments, Cell Systems (2022). DOI: 10.1016/j.cels.2022.05.004
Provided by California Institute of Technology