Study pins down role of chance in emergence of antibiotic resistance
Researchers from Konstantin Severinov's laboratory have shown what luck has to do with bacterial cells acquiring and retaining small DNA molecules called plasmids in spite of the countering action of the CRISPR-Cas defense. Genes carried on plasmids provide bacteria with resistance to antibiotics—a phenomenon plaguing people around the world. The findings provide insights on how to effectively deal with the spread of antibiotic resistance by harnessing the cellular defense system called CRISPR-Cas. The paper is published in the Proceedings of the National Academy of Sciences.
Plasmids are small circular DNA molecules. When entering a bacterial cell, plasmids tap into its resources to replicate and get transferred to daughter cells. Bacteria can fight plasmids using the CRISPR-Cas immunity system.
The CRISPR-Cas immunity comprises two key components. CRISPR is essentially a database of short fragments of foreign DNA which a bacterium has encountered before and is using to identify new invaders, such as viruses or plasmids. The "Cas" part consists of proteins that destroy foreign DNA matching CRISPR.
It was previously assumed that, once recognized by CRISPR-Cas, plasmids are either very quickly destroyed or acquire mutations that make them invisible to CRISPR-Cas.
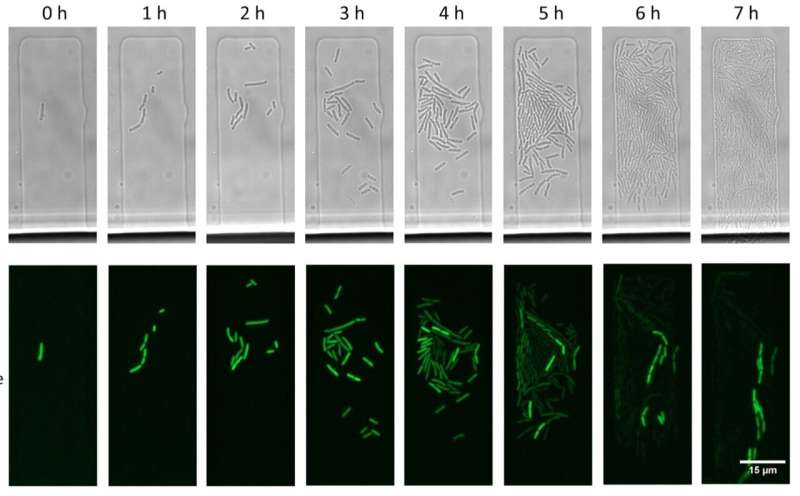
"We have proved that this is not always the case," says Skoltech Ph.D. student Viktor Mamontov, the study's lead author. "The thing is that CRISPR-Cas can take out a finite number of plasmid molecules per unit of time. Since plasmid DNA is constantly being copied, for each unique combination comprised by a certain plasmid and the CRISPR-Cas targeting it, there is a point where the rates of these two opposing processes balance each other. If such an equilibrium is reached, a plasmid can stay in the cell for extended periods of time without acquiring any mutations even in the presence of counteracting CRISPR-Cas immunity."
The team performed mathematical modeling to show that there is always a small percentage of cells in a bacterial population that manage to reach this equilibrium.
"The chances that a single plasmid molecule entering a cell will evade the CRISPR-Cas defenses are tiny. Yet, if it is 'lucky,' it can replicate and increase its numbers before it is destroyed by Cas. This not only enables plasmids to persist in cells with CRISPR-Cas but also buys them time to acquire mutations that make the plasmid fully resistant to CRISPR-Cas," Mamontov adds.
"Zooming out from plasmid survival within a single cell to processes that occur on the scale of a cell population—for example, in a bacterial colony growing on a Petri dish—we discovered another unexpected effect," Mamontov continues. "One would think that all cells in a colony grown on a medium with antibiotics will contain plasmids carrying antibiotic resistance genes even though there is a CRISPR-Cas system targeting the plasmid. We discovered this was not the case: A vast majority of cells bore no plasmids. They managed to survive thanks to a handful of cells present in the colony that achieved and maintained a balance between the rates of plasmid copying and their destruction by CRISPR-Cas. It was these rare cells that helped the rest of the population to survive in the presence of the antibiotic."
"Apparently, antibiotic-resistant cells help other cells by creating a safe haven around themselves, and a small number of plasmid-bearing cells are enough for the entire population to survive," Mamontov says.
According to the authors, the fact that plasmids can come into balance with the CRISPR-Cas defense makes a bacterial population more diverse and therefore more stable: At any given time, a small number of cells can carry a plasmid that may be useless for the time being but that could come in handy if the environment changes—and might even save the population from extinction.
The model proposed by the authors of the study incorporates the effects of plasmid replication in a single cell and on the cell population level. This makes it useful for those working on methods for removing antibiotic resistance plasmids from pathogenic bacterial cells. Biotech teams around the world are trying to adapt CRISPR-Cas for this purpose, hoping to do away with resistance in bacteria causing dangerous diseases. Understanding the intricate interactions between CRISPR-Cas and plasmids is absolutely crucial for developing this kind of tool.
More information: Viktor Mamontov et al, Persistence of plasmids targeted by CRISPR interference in bacterial populations, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2114905119
Journal information: Proceedings of the National Academy of Sciences
Provided by Skolkovo Institute of Science and Technology