May 18, 2022 feature
A hybrid open-top light-sheet microscope for versatile multi-scale imaging
During image analysis, researchers use light sheet microscopy of cleared tissue as a preferred method for high throughput volumetric imaging. A flexible system can provide a range of sizes, resolution and tissue-clearing protocols. In a new report now published in Nature Methods, Adam K. Glaser and a team of interdisciplinary scientists in mechanical engineering, bioengineering, and synthetic biology in the U.S. and Japan presented a new hybrid imaging system. Using the new method, the team combined non-orthogonal dual-objective and conventional open-top light-sheet microscopy for multi-scale volumetric imaging to visualize an intact, cleared mouse brain at the sub-micron scale. The team achieved high-throughput automated imaging of multiple specimens, and compared the outcomes with existing light-sheet microscopy systems to show a unique combination of versatility and performance in the hybrid setup.
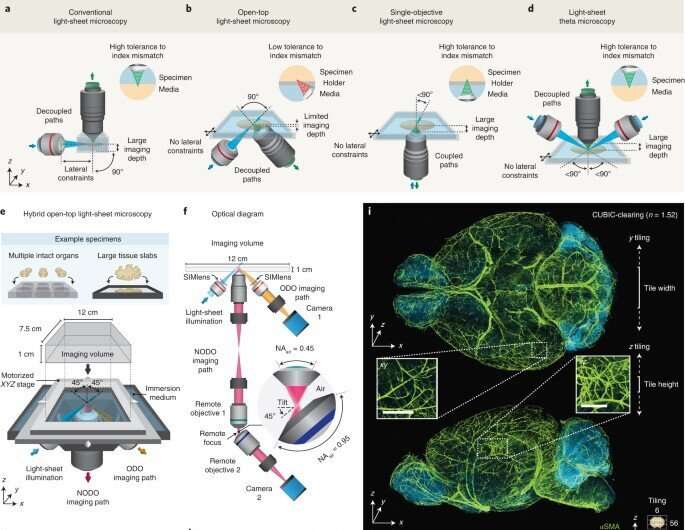
Open-top light-sheet microscopy (OTLS)
Tissue clearing protocols aim to reduce optical scattering, aberrations and background fluorescence for deep-tissue imaging with high resolution and contrast. The approach is used in many fields including neuroscience, developmental biology and anatomic pathology. The new method is a high-resolution volumetric technique to image cleared tissues with unrivaled speed and low photobleaching. Academic researchers and commercial entities have previously developed a diverse array of light-sheet microscopes; however, most light-sheet microscopes at present cannot satisfy all requirements of user-friendly standards, compatibility across protocols, large imaging depth and multi-scale imaging capabilities at the sub-micron scale. To address these shortcomings, Glaser et al developed an orthogonal dual-objective (ODO) system with tilted objectives. The open-top light-sheet microscopy (OTLS) method provided an easy technique alongside potential accessory devices including microfluidics, electrophysiology and microdissection. The team implemented a single-objective architecture and formed a hybrid open-top light-sheet microscope to open the door for advanced experiments at the sub-micron scale.
System design
Glaser et al designed the new system to feature three main objectives, which they positioned below the specimen to provide an unobstructed open top for three-dimensional imaging. The researchers included all three objectives into a monolithic imaging chamber via direct immersion and used a solid immersion meniscus lens to expand the range of refractive indices. They then developed a new non-orthogonal dual-objective light-sheet configuration alongside remote refocusing to rectify the imaging plane. The new architecture provided superior resolution to that of a single-objective light-sheet system, with broader applications for cleared-tissue imaging. The team then used the hybrid system in two example studies to represent multiscale imaging enabled time- and data-efficient experimental workflows.
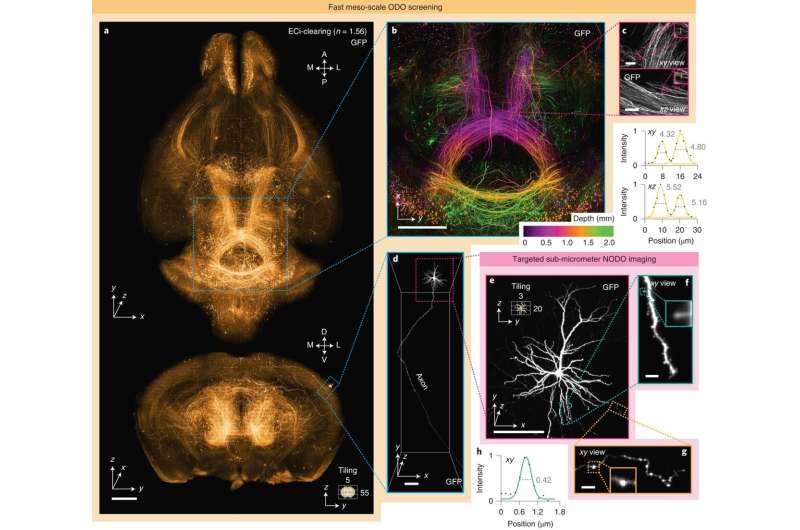
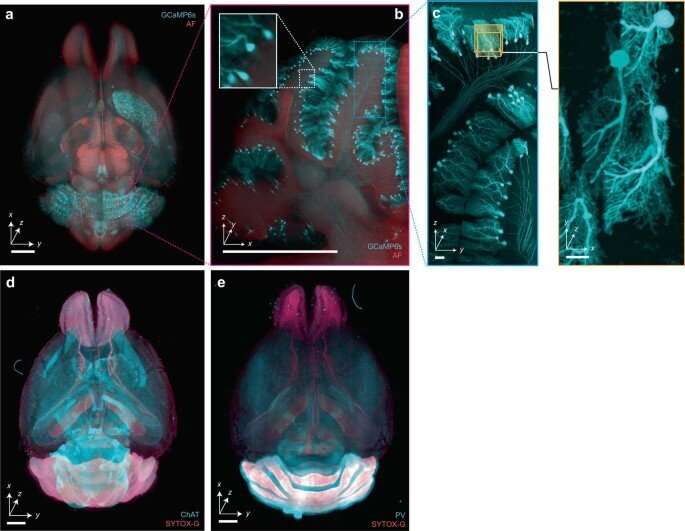
Targeted imaging: Sparse axons
During the first experiment, Glaser et al imaged axons in an intact mouse brain. Tracking the axons of individual neurons presented a challenging problem, since axons are very thin and span very large distances, the researchers therefore relied on bright labeling of a few neurons with high resolution and high contrast imaging to capture the entire brain. The scientists generated large datasets of sub-micron level imaging for computationally intensive downstream pipelines for data handling, storage and processing. Using the hybrid multiscale imaging system, Glaser et al simplified the process to screen an entire mouse brain at low resolution and identify target regions of neurons, followed by high-resolution imaging, to categorize a specific region of interest.
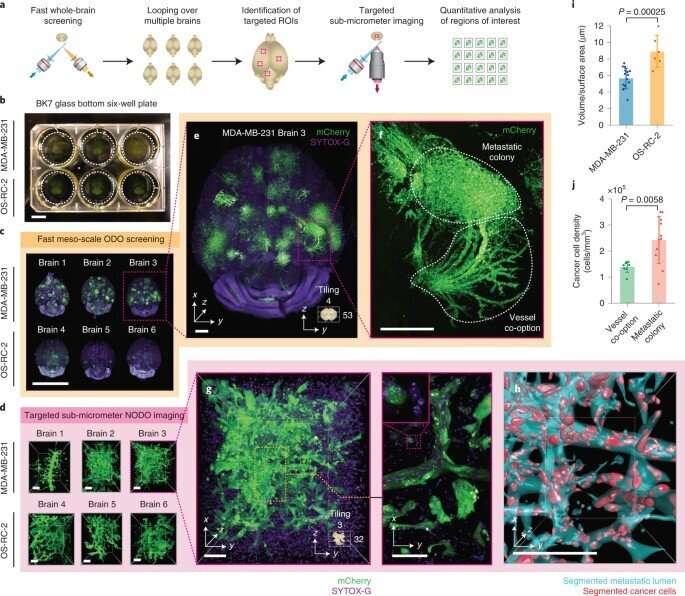
Multiscale imaging metastatic colonies
During further experiments, the researchers performed broad multiscale imaging across multiple specimens. For this, they used the hybrid OTLS (open-top light-sheet microscopy) system, and studied metastatic colonies from two cancer cell lines within intact mouse brains. Typically, it is difficult to identify the sparse and unpredictable spatial distribution of brain metastases in whole brains due to the absence of a low resolution screening method. Glaser et al therefore first identified the metastatic sites and then aimed to conduct high-resolution quantitative analysis of these regions thereafter. While studies in the past have relied on the individual manual transfer of specimens between different microscopic systems, in this case the team analyzed six intact mouse brains via the hybrid system during a single imaging session. The hybrid method reduced overall hardware costs to simplify multiscale datasets and allowed automation of multiscale imaging experiments to prevent the need to manually transfer specimens between different imaging systems. For example, while previous research required two separate microscopic systems, and weeks of tedious imaging, in addition to manual specimen transfer, the hybrid OTLS system allowed the entire experimental imaging process to be completed in two days.
Additional experiments
Aside from the experiments conducted as proof of concept, the device is also suited for an additional array of imaging studies. The team highlighted the role of the system in several projects, including multi-scale non-destructive 3D pathology of prostate cancer, whole-brain imaging of endogenous fluorescent proteins, 3D imaging of mouse embryos, imaging plants and pigmented animal models as well as large-scale imaging of human brain slices. As with all light-sheet microscopy methods, the researchers highlighted various factors underlying the process, including system design, sample preparation and sample mounting that contributed to imaging quality.
Outlook
In this way, Adam K. Glaser and colleagues showed how axial and lateral resolution of an imaging system can be improved via a hybrid open-top light-sheet microscopy (OTLS) system. In the present setup, the team chose three separate objectives; however, they point out that it is also possible in theory to achieve improved performance with an optimized non-orthogonal dual-objective, or with a single high numerical aperture with a large field of view and working distance. In summary, the hybrid OTLS experimental setup provided a practical platform with commercially available optical components to achieve an impressive balance of performance and versatility for cleared tissue imaging experiments. The team also envision the possibility of integrating high-speed voice coil actuators and tunable mirrors as alternate devices integrated to the system in the future.
More information: Adam K. Glaser et al, A hybrid open-top light-sheet microscope for versatile multi-scale imaging of cleared tissues, Nature Methods (2022). DOI: 10.1038/s41592-022-01468-5
Journal information: Nature Methods
© 2022 Science X Network