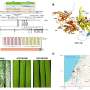
The photo shows a maize plant infected by the fungus. Credit: Khong-Sam Chia
Plant colonization by biotrophic pathogens requires sophisticated strategies for tissue invasion, defense suppression and metabolic manipulation to loot nutrients necessary for their growth and reproduction. The biotrophic fungus Ustilago maydis secretes a multifaceted effectome during maize plant colonization to achieve the abovementioned tasks. Effectors are microbial secreted manipulative molecules that act outside or inside the host cell. They are a type of microbial "weapon" that attack and trick the host's immune system to drive infection.
Co-evolutionary forces on the effectome and the corresponding host immune system shape plant-pathogen interactions and are summarized in the famous ZigZag model, where pathogen recognition by host immune receptors and recognition suppression by new effectors lead to compatible or incompatible interactions over the evolutionary lifespan of this interaction. The plant reacts to the pathogen with a first response, the pathogen triggered immunity (PTI). With help of the effectors, however, the pathogen can succeed in overcoming this first line of defense. The plant then reacts with a second response, the effector triggered immunity (ETI).
One commonality in PTI and ETI defense is the rapid formation of reactive oxygen species (ROS)—such as hydrogen peroxide—in the apoplastic space of infected plant cells. The ROS interfering protein 1 (Rip1), in turn, is an effector that suppresses this reaction. "In our study, we functionally characterize the ROS-burst-interfering effector protein Rip1 of U. maydis and provide evidence that it suppresses apoplastic ROS formation by acting in various subcellular compartments of the host cell," explains Prof. Dr. Armin Djamei from the Institute of Crop Science and Resource Conservation (INRES) at the University of Bonn and former head of the research group Biotrophy and Immunity at IPK Leibniz Institute.
"Importantly, we provide biochemical and genetic evidence that Rip1 targets and binds Zmlox3—a maize gene from the lipoxygenase family—to suppress PTI and Rip1-mediated reduction of susceptibility of maize to U. maydis."
A study on this maize susceptibility factor in the context of U. maydis infection had been recently published as a result of a collaborative work between the Kumlehn group and Djamei laboratory (Pathi et al. 2021). The newer study is published in The Plant Cell journal. Whereas in the previous work genetic evidence was provided showing the relevance of the host target Zmlox3 for U. maydis infection, here Rip1 has been identified as a corresponding effector targeting Zmlox3. On the mechanistic side, the researchers show that Rip1 relocates Zmlox3 to the nucleus and that nuclear Zmlox3 independent of its enzymatic activity leads to a reduced ROS-burst responsiveness in plants, explaining partially the ROS-burst suppressive activities of Rip1.
The complexity as a result of co-evolution gets revealed looking at the genetic data of the recent publication. Maize mutants in which Zmlox3 was knocked out showed higher resistance to infection. When the same maize mutants were infected with U. maydis strains lacking Rip1, the plants were susceptible to maize wildtype levels again. "This suggests that Rip1 triggers resistance mechanisms in the plant which are suppressed in the presence of Zmlox3 and via the action of Rip1," explains Dr. Indira Saado, first author of the study, who started her research on this topic at IPK. "Our findings indicate that the activity of Zmlox3 as a susceptibility factor is directly linked to the presence of Rip1 on the fungal side."
Together, the results reveal that the U. maydis effector Rip1 acts in several subcellular compartments as suppressor and that targeting and binding of Zmlox3 by Rip1 is responsible for the suppression of Rip1-dependent reduced susceptibility of maizeto U. maydis.
More information: Accepted manuscript, Effector-mediated plant lipoxygenase protein relocalisation triggers susceptibility, The Plant Cell (2022). DOI: 10.1093/plcell/koac105
Provided by Leibniz Institute of Plant Genetics and Crop Plant Research