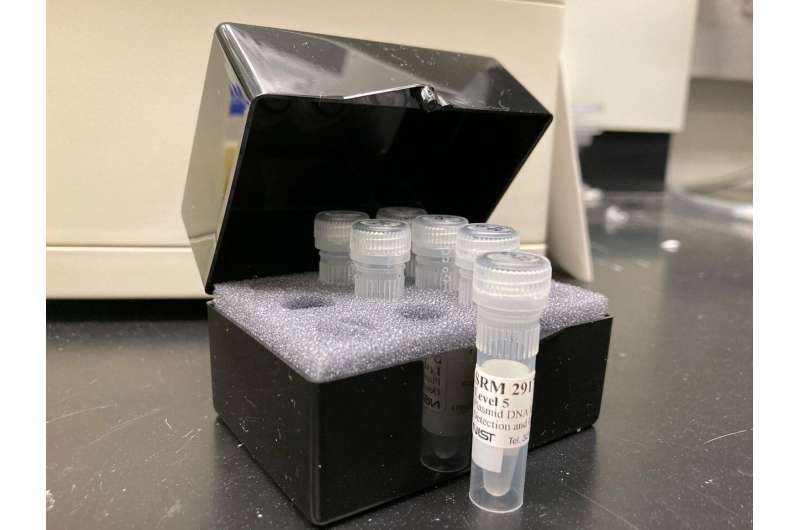
SRM 2917, Plasmid DNA for EPA-Developed Fecal Indicators, is a standardized reference material that contains 13 genetic markers, known DNA sequences, that can be used to identify a pollution source or estimate the concentration of fecal pollution in a sample. Credit: J. Kralj/NIST
As we approach summer beach season, you may occasionally encounter a "keep out" sign indicating that the area is temporarily closed. If fecal pollution is suspected, public health officials can run lab tests on water samples to check whether or not it's safe for people to swim in the water.
Despite the many lab methods available for health officials, the results are not always consistent. To help solve this issue, the National Institute of Standards and Technology (NIST) has collaborated with the Environmental Protection Agency (EPA) to develop a reference material—known as SRM 2917 Plasmid DNA for EPA-Developed Fecal Indicators—to help ensure more accurate results across the board for the methods public health officials are using.
"The key objectives for the development of NIST SRM 2917 were to help standardize the use of water quality monitoring methods across the nation and to develop a tool that can be used to establish method performance metrics," said EPA research geneticist Orin Shanks, who worked on the development of the SRM with NIST researchers. "The SRM can also help scientists, managers and the public evaluate the quality of measurements."
The EPA is responsible for developing tools to monitor recreational water quality, which includes beaches, lakes and rivers used for fishing, boating, swimming, surfing and other purposes. Fecal contamination of these waters could result from faulty septic systems, leaky sewer lines, stormwater discharges, agriculture runoff, overflows from combined sewer systems, wildlife or improper disposal of pet waste, as well as illegal activities, said Shanks.
People exposed to fecal contamination, whether through skin contact or swallowing the water, can suffer anything from minor illnesses to potentially fatal diseases. Children, the elderly and those with compromised immune systems may be at a higher risk.
Designed to help mitigate this issue, NIST SRM 2917 contains 13 genetic markers—known DNA sequences—that can be used to identify a pollution source or estimate the concentration of fecal pollution in a sample. The SRM was constructed from a plasmid, a small, often circular DNA molecule found in bacteria and other cells.
The EPA selected the SRM's genetic markers, some of which rely on over a decade of collective research. The genetic markers on the plasmid DNA serve as indicators of fecal pollution. "Fecal indicators are targets such as bacteria or genetic markers that signal there is fecal contamination," said NIST researcher Scott Jackson. "There are a total of 13 genetic markers, some of which detect human feces, cow, bird and dog feces. So, we can differentiate which fecal sources are present in a polluted water sample."
When putting together a standard reference material (SRM), researchers use what they call the "robottler," a robotic machine they've programmed to automatically fill test tubes. SRM 2917 (Plasmid DNA for EPA-Developed Fecal Indicators) consists of 13 known DNA sequences (genetic markers) that can be used to estimate the concentration of fecal pollution in a sample. Here, the test tubes for the SRM are being filled with the plasmid DNA sample. Credit: J. Kralj/NIST
Each genetic marker of the SRM functions with a specific qPCR method designed to characterize fecal pollution in a water sample. A qPCR assay—quantitative polymerase chain reaction—is a molecular test used to estimate a sample target DNA concentration that relies on a reference material to interpret results. Three of these testing methods are nationally validated by the EPA.
The qPCR methods target two types of fecal indicators: The first estimates the total amount of fecal matter in a sample, and the second identifies the source of the fecal contamination. Two qPCR methods target fecal indicator bacteria, E. coli and Enterococcus, which are found in the fecal waste of most mammals. The remaining 11 qPCR methods help identify sources such as human, cattle, dog, pig and bird fecal matter.
"This is a multiuse reference material. A future lab can use this SRM to help determine if water is safe to swim in and characterize what the sources are when there is fecal pollution," said Shanks.
What makes the SRM so novel is that all 13 genetic markers are on one physical piece of DNA, said NIST researcher Jason Kralj. This means that researchers can assess multiple qPCR method results from a water sample using one standard.
The SRM could be used to help minimize variability in water quality measurements between labs and assist in the process of lab accreditation, which indicates that a public health lab is running its tests correctly. Even though the SRM was developed for routine recreational water quality testing, it can also be used to support stormwater management, food production monitoring, wastewater surveillance and outbreak exposure route identification, said Shanks.
A national interlaboratory study was conducted during the development of the SRM. In the near future, NIST and the EPA will release the results of the study, which was designed to assess the performance of the SRM on a national scale.
"Looking down the road, it is likely that the success of this collaboration will lead to future collaborations focused on the development of new reference materials that support the implementation of methodologies designed to protect environmental health and natural resources," said Shanks.
Provided by National Institute of Standards and Technology