Hundreds of gene regulatory motifs cooperate and conflict to make brain cells
DNA is considered the blueprint of our human bodies. It codes for the tens of thousands of genes that determine whether a cell transforms into a brain cell, heart cell or even cancer cell. Other DNA sequences, known as enhancers, determine if genes are activated or inactivated. In a new study seen in Nature Communications, ASHBi Associate Professor Fumitaka Inoue and colleagues in the United States report the "massively parallel reporter perturbation assay" to study how enhancers are fine-tuned over time during the generation of brain cells.
Enhancers determine when, where and to what degree a gene is turned on. Enhancers themselves are regulated by transcription factors, which target regulatory motifs in the enhancer sequence. Thus, interactions between enhancers and transcription factors are critical for enhancing or suppressing the genes that determine a cell's fate and behavior.
"Enhancers have important effects on gene expression and thus determine whether cells will grow into different organs. Despite their consequences, however, we understand relatively little about how enhancers function," explains Inoue.
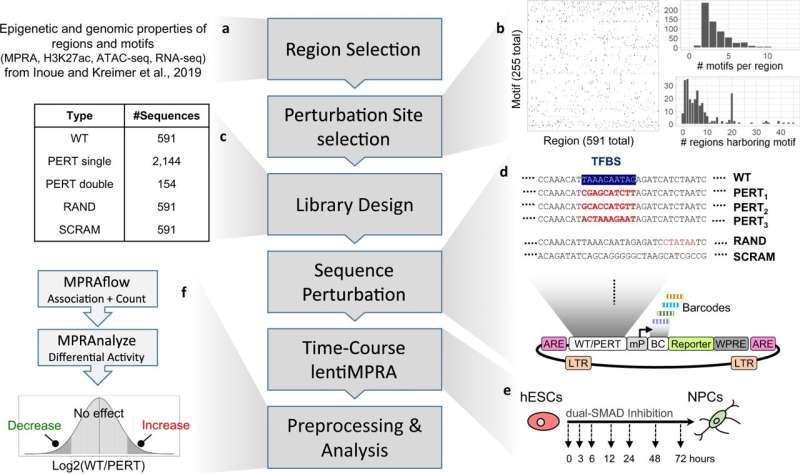
A number of biochemical assays are available to study enhancers, but massively parallel reporter assays (MPRA) allow for the quantitative analysis of thousands of enhancers at one time.
"MPRA are useful for studying the interplay between enhancers and transcription factors. To extract even more information from the assays, we modified the assays to introduce perturbations in the regulatory motifs where transcription factors bind," continues Inoue.
These perturbations were introduced in multiple regulatory motifs in thousands of enhancer sequences. MPRA was then applied to study the effects of the perturbations on the enhancer activity and thus systematically identify which regulatory motifs are important for gene activation or inactivation.
To validate their massively parallel reporter perturbation assay, the team applied it to the differentiation of human embryonic stem cells into neural cells. They found 598 regulatory motifs that either enhanced or dampened transcription.
"We broke these motifs into four groups. Essential and contributing motifs either were required or contributed to the activation. Likewise, silencing and inhibiting motifs were required or contributed to dampening transcription," says Inoue.
Further investigation revealed that among the genes enhanced or dampened, many are involved in promoting neural differentiation or stem cell pluripotency. Moreover, the assay clarified that in most cases the regulatory motifs determined whether transcription was enhanced or dampened, but the timing of the perturbation determined the magnitude of the effect. Overall, says Inoue, the findings demonstrate an intricate network of regulatory sequences that interact with one another to achieve neural differentiation.
"The biggest accomplishment of the assay is the number of sequences we could analyze and perturbations we could apply. As a result, we demonstrated the complexity of many models for how transcription factors regulate gene expressions," he said.
More information: Anat Kreimer et al, Massively parallel reporter perturbation assays uncover temporal regulatory architecture during neural differentiation, Nature Communications (2022). DOI: 10.1038/s41467-022-28659-0
Journal information: Nature Communications
Provided by Kyoto University