Genomic regulatory map of the zebrafish
Zebrafish and humans look very different on the outside. Yet about 70 percent of their genes are similar to human genes—including many that can trigger diseases. That makes the animal a popular model organism. Many observations of biological processes such as embryonic development can be transferred to humans. The vast majority of genes that play a role in this process have been identified.
The situation is different for sequence segments within the DNA molecules that regulate the corresponding gene. "In a sense these are switches that activate the gene at the right time and at the right place, or via an appropriate signal," explains Professor Uwe Ohler of the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), who leads a lab at the MDC's Berlin Institute for Medical Systems Biology (BIMSB). He compares gene regulation to the lighting in a big house that has been elaborately wired. "You don't know, for example, which switch needs to be pressed to turn on the basement light. Cells are wired in complex ways too." This means, he says, you have to figure out which switches are present in which cell type, which of them are important, and what they regulate. But concrete details about their occurrence as well as their function and structure are often lacking.
In the paper appearing in Cell Genomics, Ohler, along with study leader Dr. Scott Allen Lacadie and other colleagues, is now presenting a comprehensive genomic regulatory map of a 24-hour-old zebrafish embryo. First authors Dr. Alison McGarvey and Dr. Dubravka Vučićević used single-cell technologies to examine the genome of approximately 23,000 nuclei from the entire fish organism. Self-learning algorithms then analyzed and categorized the data sets to determine which switches are active in which cell type.
"A switch that turns on a gene in liver cells looks different from one that is responsible for a gene in brain cells," Ohler says. "This is the only way the organism can activate certain switches individually and at the right time." These regulatory segments control things like gene transcription. First, they are transcribed into a transportable form (mRNA) so that vital proteins can be produced in subsequent steps.
Genomic regulatory map: putting the puzzle together
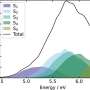
There are, Ohler says, millions of such switches in a typical vertebrate genome—and somewhere between 50,000 and 100,000 are active in each cell. The experiment devised by the team can identify only switches that are activated in the cells, which is done based on the structure of the chromosomes in the genome. These open up slightly, thereby enabling the right proteins to bind to them and activate the switch. "On average we can identify about five to ten thousand switches in a single cell using this method—perhaps five to ten percent of the total number per cell," Lacadie says. "And that in turn is only successful for a certain percentage of all cells in the embryo." Yet to obtain relatively complete information about the entirety of regulatory genomic segments in a cell type, the researchers had to properly combine the data from the many thousands of single cells analyzed.
To accomplish this requires computer programs that sort the cells according to their tissue of origin, such as brain or muscle. That sounds easy enough, but it is actually a huge challenge because each cell's data set is like a puzzle piece that has to be matched to an unknown puzzle. "But some of the regulatory switches we discover in one single cell overlap with those we find in another cell," Lacadie explains. In other words, pieces from the same corner of the puzzle share commonalities. And these in turn "serve as clues that help our computer programs to group the analyzed cells into cell types," Lacadie says.
The machine learning algorithms used for this task come from artificial intelligence, which learns on its own and continuously improves with time. The programs were developed under the leadership of Dr. Wolfgang Kopp, another first author of the study. It is very difficult to say how many of the switches that actually exist in zebrafish have ultimately been included in the genomic regulatory map, according to Lacadie, who adds: "We're maybe in the 80 percent range for muscle and brain tissue, where the most cells were characterized, but we're well below that for other cell types."
So while there is still much work to be done, the results are a big step forward for zebrafish research specifically and for the study of gene regulation during development generally, Lacadie sums up. The team's data are now being incorporated into a collaborative project in which several other labs are contributing genomic data to create a comprehensive mapping of the zebrafish genome across the different stages of development.
Biased data are no problem
Unlike previous computational analysis methods, the algorithms can analyze the raw data collected in experiments without the team having to prepare them beforehand, Ohler says. This is especially crucial if the data were collected with different technical equipment or in different laboratories. As a rule these are subject to a bias, which means that the data sets are distorted and not directly comparable. To filter out the bias and thus enable the data to be combined, the lab trained the algorithms to look for those pieces of information in the data sets that betray their source. These features are then ignored. The assumption is that the information that remains is biologically relevant, Ohler says.
Kopp, Ohler, and their colleague Dr. Altuna Akalin recently described these newly developed machine learning methods, which can analyze genomic data sets from different single-cell technologies despite the presence of bias, in the journal Nature Machine Intelligence. "We now want to use the algorithms to find out which regulatory DNA sequences are relevant to genetic diseases," says Ohler. "Initially we just looked at gene mutations, but now we can also examine which switches are involved and not functioning properly." The team believes that especially for many common diseases with a genetic predisposition, answers can be found in the switch regions. So it will be exciting to see what the newly developed algorithms reveal in the future.
More information: Alison C. McGarvey et al, Single-cell-resolved dynamics of chromatin architecture delineate cell and regulatory states in zebrafish embryos, Cell Genomics (2022). DOI: 10.1016/j.xgen.2021.100083
Wolfgang Kopp et al, Simultaneous dimensionality reduction and integration for single-cell ATAC-seq data using deep learning, Nature Machine Intelligence (2022). DOI: 10.1038/s42256-022-00443
Journal information: Nature Machine Intelligence
Provided by Max Delbrück Center for Molecular Medicine