Estimating the strength of selection for new COVID-19 variants
As the discovery of the new omicron variant illustrates, new COVID-19 variants will continue to regularly emerge. In an effort to make sense of these new variants, scientists at Los Alamos National Laboratory have developed methods to quantify how much more or less transmissible they are, which could have far-reaching implications for public health in terms of COVID-19 risk and the vaccination levels required to obtain herd immunity.
"Generally, new COVID-19 variants are simply discussed in terms of being more dangerous or spreading quicker than previous strains," said Ethan Romero-Severson, a computational epidemiologist in Los Alamos's Theoretical Division and senior author on the paper published today in Nature Communications. "We showed that it is possible to calculate new strains' transmission advantage while accounting for alternative explanations such as migration and random genetic drift. Our collection of methods allows us to look both broadly at the global situation and in greater detail at specific countries using publicly available genetic sequence data."
The Los Alamos research is "a method for integrating molecular epidemiological surveillance into surveillance systems using publicly available data streams," Romero-Severson noted.
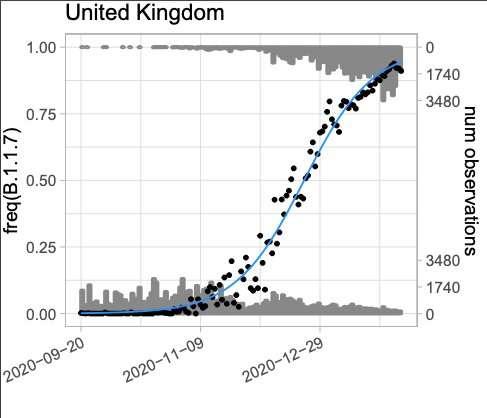
The team used two distinct but complementary approaches. The first is derived from classical population genetic methods that relate the increased transmissibility of a COVID-19 variant to the expected frequency of that variant in the population over time.
"We modified that model to include migration as a possible alternative explanation to increased transmissibility and implemented it in a hierarchical modeling framework that allowed us to estimate the unique selection effect for each variant in each country in which it appeared," he said.
The second, more detailed method used a stochastic (allowing for uncertainty) epidemiological model to predict both the changes in COVID-19 variant frequencies and deaths over time, accounting for natural and random variation in the virus both between and within countries over time.
Together, these approaches showed that the pattern of emerging and rising COVID-19 variants globally was driven by large increases in the transmissibility of the virus over time. The methods also clearly established that early detection of variants of concern is possible even when the global frequency of new variants is as small as 5 percent.
More information: Christiaan H. van Dorp et al, Estimating the strength of selection for new SARS-CoV-2 variants, Nature Communications (2021). DOI: 10.1038/s41467-021-27369-3
Journal information: Nature Communications
Provided by Los Alamos National Laboratory