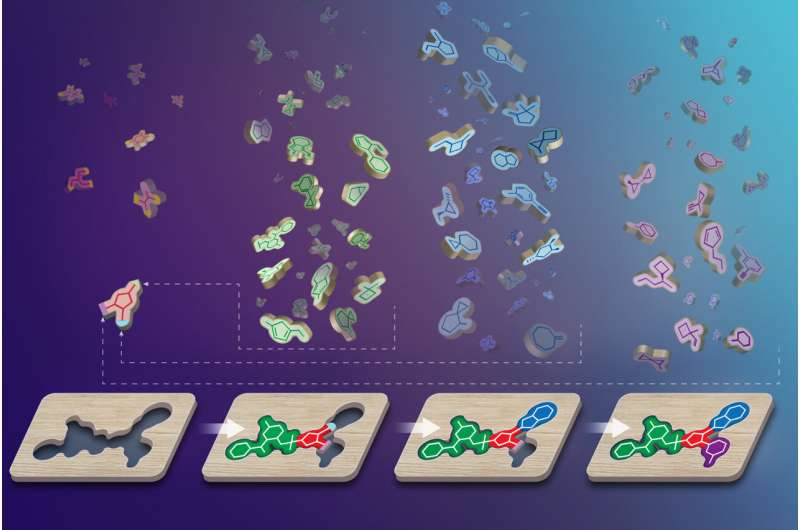
Chemists can build effective drugs using chemical components called synthons, represented by the colorful puzzle pieces, that match the shape of a therapeutic target on a protein, represented by the wooden block. Credit: USC Dornsife/Yekaterina Kadyshevskaya
On the surface of our cells are docking stations called receptors. All kinds of compounds from caffeine and dopamine to heroin, THC and LSD bind to these receptors. In fact, G protein-coupled receptors are the intended targets of more than 30 percent of pharmaceutical products currently on the market. But these drugs often hit unintended targets—think carpet bombing the nervous system—leading to the laundry list of side effects commonly heard at the end of pharmaceutical commercials.
"What we need are more precise and less harmful treatments that are just as effective," said Bryan L. Roth, MD, PhD, the Michael Hooker Distinguished Professor of Pharmacology at the UNC School of Medicine. But creating these better drugs isn't easy. Drug developers need to know the exact chemical structure of a drug and the intended receptor to produce the exact chemical reaction we want inside cells. The kicker is to make sure the drug doesn't affect other receptors or bind to the target receptors but bind in such a way to trigger unintended consequences inside cells.
"In the past, scientists would test molecules in a one-by-one fashion against a therapeutic target in a lengthy and expensive operation," said Roth, who leads the National Institute of Mental Health Psychoactive Drug Screening Program. So scientists created virtual libraries of molecules and fancy computer programs to test hundreds of thousands of molecules at a time. By "test" we mean that the computer program scoured a database of molecules that theoretically could be created and theoretically could bind with the proper affinity to a given cell receptor. And then scientists could tinker with chemical bonds in a computer program to optimize a molecule's structure. Then scientists could physically make a tiny sliver of these molecules and test them in cell cultures.
The creation of virtual libraries was a giant leap in the field. Testing hundreds of thousands of possible molecules, some of which might be candidates with properties worth investigating, sounds like a lot. But there are billions upon billions of possible chemical combinations that could theoretically lead to the creation of a near infinite number of molecules or potential drugs. As a result, scientists wound up creating vast libraries of theoretical compounds, billions of mostly "undiscovered" and unexplored molecules that may or may not bind to a given cellular target; they may or may not have therapeutic value at all.
"Unfortunately, chemical space is vast," Roth said. "It has been estimated that there exists, theoretically, more chemicals than there are actual molecules in the universe. And only a small sliver of the potential chemicals can be physically tested."
So Roth teamed up with researchers from the University of Southern California and Northeastern University to validate V-SYNTHES, a new type of computational method developed by Vsevolod Katritch, PhD, at USC that allows scientists to first identify the best combinations of chemical building blocks called synthons – hypothetical units within molecules – to serve as seeds that can grow into a hierarchy of molecules with the best predicted ability to bind to the receptor targets.
"This approach allows researchers to computationally test billions of compounds against a therapeutic target," Roth said. "To our knowledge this is the largest successful computational screen to date."
As described in their Nature paper, they tested 11 billion theoretical compounds against a cannabinoid receptor (CB2) that marijuana's active ingredient THC targets. The Katritch lab developed the approach and performed the computational studies. With help from the Northeastern Lab of Alexandros Makryannis, PhD, and Roth's lab, the researchers discovered a new compound that is a highly selective and potent CB2 antagonist – a compound that dampens or blocks the activity of a receptor. Then the Makryannis and Roth labs validated this screening and discovery. They further validated this process against another target, the kinase ROCK1, a known drug target of many health conditions.
"V-SYNTHES represents a major advance in the field of drug discovery," Roth said. "It is easily scalable and adaptable, and it should open new vistas in the discovery of potentially therapeutic chemicals for a large number of disorders at a rate never before possible."
More information: Vsevolod Katritch, Synthon-based ligand discovery in virtual libraries of over 11 billion compounds, Nature (2021). DOI: 10.1038/s41586-021-04220-9.
Journal information: Nature
Provided by University of North Carolina at Chapel Hill