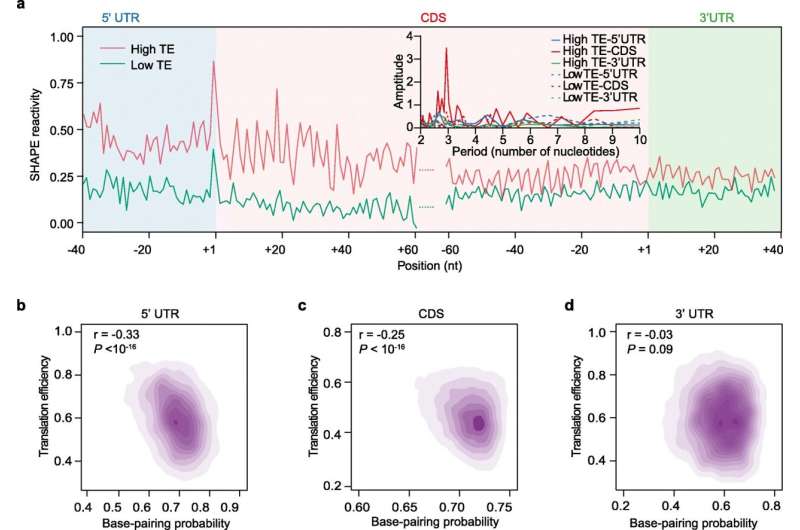
Translation efficiency (TE) correlates with in vivo RNA structures in tetraploid Kronos. a Line plot showing SHAPE reactivities of highly and lowly translated transcripts, as shown by 10% of highest or lowest TEs. SHAPE reactivities of different genic regions across highly (red) or lowly translated (bluish green) mRNAs were averaged. mRNAs were aligned by their translation start and stop codons. The inset shows strong 3nt periodicity of SHAPE reactivity in the CDS region, but not in the 5′UTR or 3′UTR of highly translated mRNAs, nor in the 5′UTR, CDS, or 3′UTR of lowly translated mRNAs. b–d Relationship between RNA structure and TE across different genic regions. The scatter plots showed the correlation between base-pairing probability and TE across 5′UTR (b), CDS (c), and 3′UTR (d). Credit: DOI: 10.1186/s13059-021-02549-y
Crops often have multiple genomes hybridized from their parents, a condition known as polyploidy.
Polyploidy is a major route to the formation of new species in plants and an important way of improving agronomic traits in crops.
In polyploids, the effect of subgenomes is not equal: the gene expression of one subgenome is dominant over the other, resulting in what is called subgenome expression asymmetry.
During the process of gene expression, DNA is transcribed into RNA, which is then used make protein, through a process termed translation. Until now it has been unclear whether there is a subgenome asymmetry at translational level in polyploid plants.
Researchers from John Innes Centre, along with collaborators in China have discovered that there is subgenome asymmetry at translational level in tetraploid wheat.
They found RNA molecular structure contributes to this translational asymmetry between the parental subgenomes. These new findings will help wheat breeders understand how traits are inherited.
Through a hybridisation and whole-genome duplication that happened around 400,000 years ago, wheat species either have two or three copies of each gene, termed as homeologous pairs.
Combining expertise in RNA biology from the Ding group with expertise of wheat genomics from the Uauy group at the John Innes Centre and the Zhang group from Northeast Normal University in China allowed the team to look at how much of the protein was made from homeologous pairs from different subgenomes.
Using the SHAPE method developed by the Ding lab to capture the snapshot of RNA structure allowed the team to study the relationship between RNA structure and translation. They found that the single strandedness of the RNA is associated with higher translation efficiency, making more proteins on these RNAs.
There are sequence variations between the homeologs, known as single nucleotide variations (SNVs). The team wanted to find out if these SNVs may cause differences in RNA structure of homeologous pairs.
They identified SNVs responsible for big differences in RNA structure, which were called "riboSNitches." The SNVs that cause RNA structure change are more subtle than other SNVs.
This research suggests that subconsciously humans over the 10,000 years of domesticating wheat may have been selecting not only through DNA sequence, but also through the structure of RNA, which has been largely unacknowledged before.
Previously plant breeding has used predictions based only on DNA sequence, but this research suggests that applying knowledge about RNA could help provide more accurate predictions of how traits are passed on.
More information: Xiaofei Yang et al, Wheat in vivo RNA structure landscape reveals a prevalent role of RNA structure in modulating translational subgenome expression asymmetry, Genome Biology (2021). DOI: 10.1186/s13059-021-02549-y
Journal information: Genome Biology
Provided by John Innes Centre