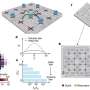
Sampling a fish farm in southern Tanzania at Songea. Credit: Bangor University
A new genomics marker tool has been shown to accurately identify tilapia species and tell apart their hybrids, providing a novel resource to help develop aquaculture and empower conservation in Tanzania, Africa.
Crucially, the new tool offers a cheaper solution than full genome data analysis—the current approach to monitoring local biodiversity.
Led by the Earlham Institute, alongside the Tanzania Fisheries Research Institute, Roehampton University, Bangor University, the University of Bristol and the University of East Anglia in the UK, the new genomics marker tool enables tilapia species identification and pinpoints hybridisation between invasive and native tilapia species.
Tropical inland aquaculture production has increased rapidly in recent decades to 47m tons in 2018. Tilapia, a group of cichlid fish dominated by the genus Oreochromis and native to Africa and the Middle East, have been a key part of this expansion - accounting for 5.5m tons of the global total. Continued expansion of inland aquaculture is particularly important in Africa, where climate change and population growth are putting vulnerable food production systems under added strain.
As a result of inland waters' temperature gradually rising due to climate change, African fisheries suffer from associated physical and chemical changes to the aquatic environment. This coupled with the continuous population growth that is estimated to reach 9 billion inhabitants in sub Saharan Africa and Asia by 2100, causes further concern for food security.
Tilapia are endemic to Africa, and East Africa including Tanzania is a hotspot of natural diversity for tilapia species. At least eight fully-endemic Oreochromis species are found in Tanzania and an additional 12 species that are endemic to catchments shared with neighboring countries. Several of these species are adapted to unique environmental conditions, such as elevated temperatures, salinity, and pH and could be of interest for future aquaculture developments.
Lead author Dr. Adam Ciezarek, Postdoctoral Scientist in the Haerty Group at the Earlham Institute, says that "one of the major difficulties in tilapia aquaculture lies with the discrimination of species. This currently relies on morphological traits, which are particularly a problem to identify in females and juveniles. This in turn can lead to accidental stock contamination. Furthermore, non-native farmed tilapia populations (Nile tilapia, O. niloticus) are known to be widely distributed across Africa and to hybridize with native Oreochromis species.
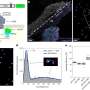
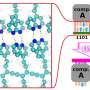
"In response to the need to accurately distinguish species but also identify potential hybrids that can be carried out cost-effectively and faster, an optimized design based on 96 single nucleotide polymorphisms (SNPs) biomarkers was developed. This design has also proven to be more accurate than microsatellite or morphological identification of interspecific hybrids.
"Tilapia are a hugely important group of fish for aquaculture. Farmed strains have been introduced to many water bodies as non-native species, which creates a problem in areas where native tilapia species are prone to hybridizing. Hybrid tilapia survive less well and grow slower, which threatens the survival of natural populations.
"This also threatens the survival of tilapia farms, as stocks are often taken from the wild. If they're stocking what they believe to be pure species, suitable for farming, but are in fact stocking slowly growing hybrids, the farm may fail.
"It's important to be able to spot these hybrids, yet it's unreliable to separate them from pure tilapia species by physical characteristics alone. We have shown that it is possible to identify them using full-genome data. Crucially, we have also demonstrated that a vastly reduced set of 96 SNPs can perform just as well—with greater efficiency and precision, and at a much lower cost."
Lead co-author Prof George Turner, School of Natural Sciences at Bangor University, says that "case studies indicate several locations where introduced aquaculture species have become established in the wild, threatening native Oreochromis species in Tanzania. At present, native Oreochromis species are poorly characterized, and their conservation could benefit from the identification of purebred populations for protection.
"Such safeguarding of the wild relatives of farmed species would also protect unique genetic resources that could be used to enhance traits in cultured Oreochromis cichlid strains."
Dr. Ciezarek added that "the novel SNP markers represent an important resource for assessing broodstock purity in fishery hatcheries, helping to conserve unique endemic biodiversity. We hope that this more affordable and convenient tool will be used to accurately assess potential farm stocks, as well as surveying natural water bodies for evidence of hybridisation between tilapia."
The paper is published in Aquaculture.
More information: A. Ciezarek et al, Whole genome resequencing data enables a targeted SNP panel for conservation and aquaculture of Oreochromis cichlid fishes, Aquaculture (2021). DOI: 10.1016/j.aquaculture.2021.737637
Provided by Earlham Institute