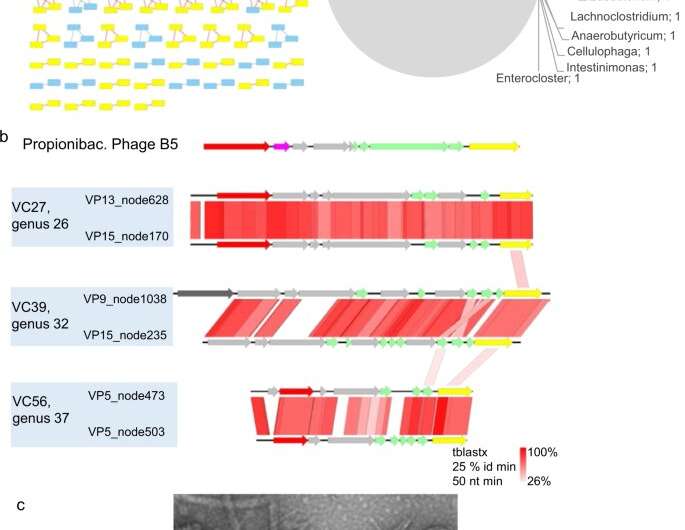
Properties of circular pig inoviruses. a, left VContact2 map of all complete genomes that could be clustered. Those encoding a gpI homologous to the one of Propionibacterium phage B5 are highlighted in yellow; a, right, host predicted for all circular genomes. b Genome alignments (tBLASTx) and Easyfig map of several inoviruses encoding a B5-like gpI gene. Yellow: gene encoding morphogenesis protein I, red: rep gene for replication initiation, green: structural genes (with transmembrane domains), pink: ssb (single-strand DNA binding) gene. Gray: gene of unknown function. Dark gray: putative phosphoadenosine phosphosulfate reductase. c Transmission electron microscope image of virome VP17, the white arrows point to a thin filament that may correspond to an inovirus (length> 420 nm, width, 8 nm). Credit: DOI: 10.1038/s43705-021-00054-8
It is a fact that bacteria are increasingly developing antibiotic resistance. Information campaigns have raised awareness among the general public that the main cause of antibiotic resistance is the high 'selection pressure' on bacteria due to massive and inappropriate use of antibiotics in both humans and animals.
But this is not the only explanation. A set of mechanisms in bacteria, known as horizontal gene transfers, are also involved. Bacteria are able to exchange DNA material with each other. Some of the DNA material that is exchanged does not contribute anything specific, while some could confer antibiotic resistance. Understanding the mechanisms by which the material carrying resistance genes is transferred could help scientists identify a new way to fight resistant bacteria.
This DNA material can either be released into the environment (the material is then free for all other bacteria to incorporate), or passed on via direct contact between two bacteria, a process called conjugation. A third route of gene transmission uses bacteriophages, or small viruses in bacteria, which are able to pass their DNA from one bacterium to another.
Bacteriophages: A vector of resistance?
Over the last decade, a question has divided the scientific community: are bacteriophages, especially those used in phage therapy, capable of transferring antibiotic resistance genes to bacteria? Although a 2013 scientific article published in Nature found that bacteriophages could indeed do so, this answer has not put the matter to rest.
The controversy caught the attention of a team of INRAE researchers specialized in bacteriophages. The scientists conducted metagenomic research (analysis of the whole genome) on bacteriophages from pig fecal samples of 14 farms. Using in-depth computer analyses, the researchers finely characterized the DNA of all these bacteriophages and came to a clear conclusion: the bacteriophages studied do not possess a single antibiotic resistance gene. They cannot therefore supply resistance genes to bacteria.
This research sheds light on the issue of antibiotic resistance, a key public health concern. Bacteriophages continue to be used as allies in the fight against antibiotic-resistant bacteria through phage therapy. This practice, which is not yet widespread, uses bacteriophages on patients infected with bacteria that are resistant to all antibiotics. Bacteriophages attack and destroy the resistant bacteria, without the possibility of passing on resistance genes to other bacteria. The therapy offers hope in the fight against the growing phenomenon of antibiotic resistance.
More information: Maud Billaud et al, Analysis of viromes and microbiomes from pig fecal samples reveals that phages and prophages rarely carry antibiotic resistance genes, ISME Communications (2021). DOI: 10.1038/s43705-021-00054-8
Journal information: Nature
Provided by INRAE