November 30, 2021 feature
Mapping enzyme catalysis with metabolic sensing
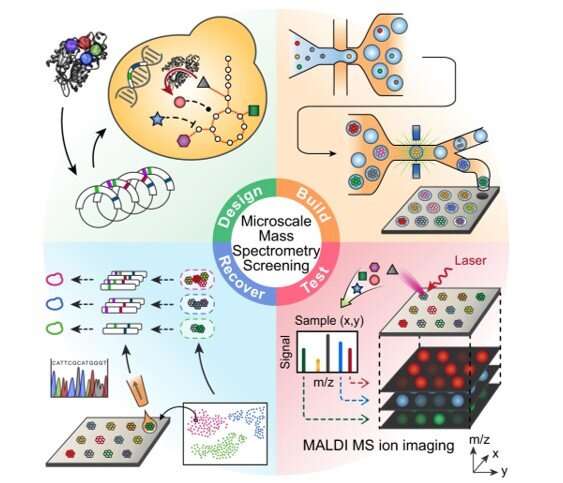
Enzymes maintain a range of protein sequences and diverse structural forms with activities that far exceed the best chemical catalysts. However, research on engineering them with new and improved features are limited due to technical inadequacies. In a new report now published in Nature Communications, Linfeng Xu and a research team in bioengineering, chemistry, and molecular biology at the University of California, the University of Michigan, and the University of Texas, in the U.S., described a scalable approach to characterize enzyme activity using the metabolism of the host cell as a biosensor to understand product formation. The team then measured the metabolic pathways of different products via mass spectrometry and the results provided them a general path to sense product formation and discover unexpected results and map the effects of mutagenesis.
Metabolic engineering: Designing enzymes in the lab
Enzyme engineering is based on an iterative cycle, where libraries of gene variants can be designed and synthesized into proteins to test for the activity of interest. While research efforts have shown success, most tests remain at a bottleneck. The dominant strategy of the research work is based on well plate screening to directly measure the product of the enzyme. However, this method is limited in scalability since it could only test just hundreds of variants per cycle. Alternative approaches include flow cytometry and droplet microfluidics for selections, but they are constrained by an inability to directly detect product formation. To enhance the ability to engineer enzymes through screening, a new method is appropriate, wherein the generality of well plates can be combined with the scalability of microfluidics. In this study, Xu et al. described a screening approach that combined the scalability of microfluidics with the generalization of mass spectrometry.
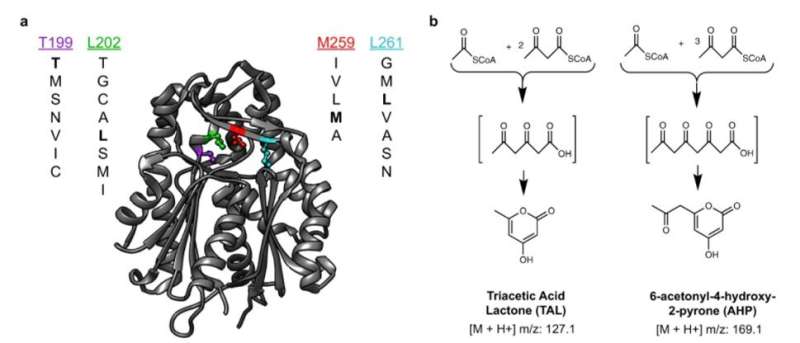
Metabolic engineering: Mechanism of action
During this approach, the team used the background metabolism of the host as a biosensor to assess the activity of an enzyme embedded therein and used micro-scale mass spectrometry technology to characterize the metabolic changes. The enzyme catalyzed the molecules of central metabolism and perturbed or disturbed the metabolite profile of the host to generate a signature. Xu et al. then measured this signature, even if the enzyme's product was not directly measured. This approach provided a general method to map the catalysis of a mutated enzyme to understand the range of products that could be generated and then recover the sequences of variants with desired activities or characteristics. During the work, Xu et al. used matrix-assisted laser desorption ionization (MALDI)-mass spectrometry for single-cell metabolomics, and also to identify enzyme products from microbial colonies. The team then combined the approach with printed droplet microfluidics (PDM) to prepare, print and screen all mutants from a library of Gerbera hybrida, plant-specific polyketide synthases. The enzyme was responsible for the biosynthesis of triacetic acid lactone (TAL) via the condensation of a starter acetyl-CoA unit with two malonyl-CoA molecules. Researchers had also used TAL as a platform precursor to synthesize high-value chemicals derived from fossil fuels, in which any mutations of active sites in such enzymes can alter the kinetics and spectrum of products. Culture expansion during the study produced additional materials when compared to a single cell, providing a signal boost with accurate metabolic data.
Mapping the catalytic activity of an enzyme variant library
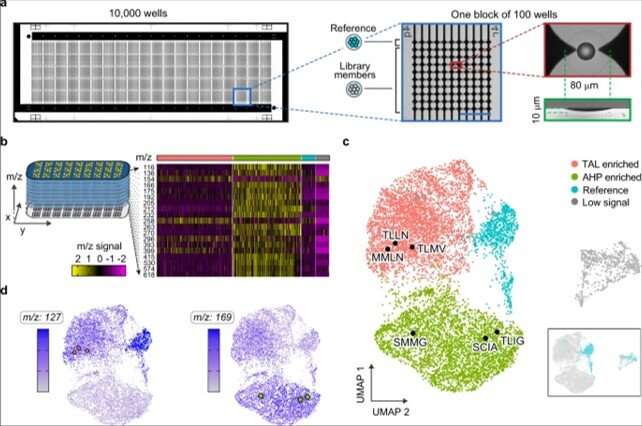
To conduct microscale mass spectrometry, Xu et al. used a high-density plate with a glass slide etched with 10,000 wells to concentrate the material to the center for accurate quantitation. They loaded all wells with one colony to accomplish PDM (printed droplet microfluidics) in 30 minutes. During printing, the team scanned the colonies and printed 9000 library members and 1000 reference strains. They dried the plate and prepared it for matrix-assisted laser desorption ionization (MALDI)-mass spectrometry imaging to provide data on all detectable metabolites via mass-to-charge ratio. Most importantly, the method inherently possessed soft ionization to allow polymerase chain reaction (PCR)-based recovery of enzyme genes after analysis.
Detecting the catalytic activity of enzyme variants with metabolic biosensing
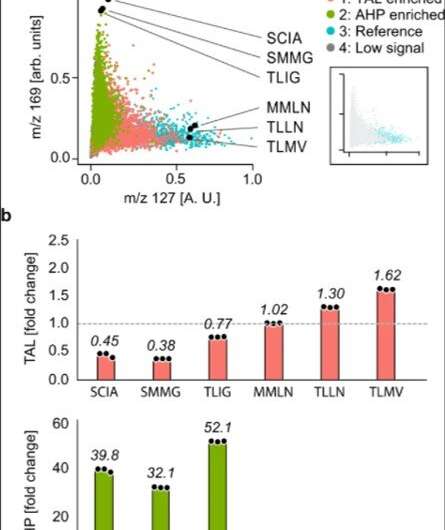
The researchers used the method to extract information on many molecules in the host cells in order to discover unexpected enzyme activities. They plotted alterations in the cell metabolome as a uniform, manifold approximation and projection (UMAP) to obtain high dimensional data while preserving cluster information. Using the data, Xu et al. observed four clusters corresponding to metabolite profiles. The outcome highlighted two major activities that distinctly perturbed cell metabolism. To characterize how these activities related to the enzyme sequences, the team selected and sequenced mutants from each cluster. As a result, the host cell's metabolite profile provided a biosensor to detect changes in the embedded enzyme's activity.
Outlook
In this way, Linfeng Xu and colleagues showed how metabolite sensing allowed them to detect product formation without directly detecting the products, as accounted using enzyme activity, which perturbed the metabolite profile of the host cell. Given that such perturbations are distinct for each activity, they detected multiple activities generated by a mutant library. Such indirect sensing is beneficial for enzyme engineering. In addition to providing a universal readout of product formation, the team explored the analytic approach with cells and matrix-assisted laser desorption ionization (MALDI)-mass spectrometry methods with applications relevant to other bioproduction systems. This approach can apply to any engineering method that perturbs host cell metabolism, including genetic circuits and biosynthetic pathways.
More information: Linfeng Xu et al, Mapping enzyme catalysis with metabolic biosensing, Nature Communications (2021). DOI: 10.1038/s41467-021-27185-9
Etienne Becht et al, Dimensionality reduction for visualizing single-cell data using UMAP, Nature Biotechnology (2018). DOI: 10.1038/nbt.4314
Journal information: Nature Communications , Nature Biotechnology
© 2021 Science X Network