Enlarging windows into understanding gene functions

In a text file, the rows of letters A, T, C and G appearing over and over in a dizzying array of combinations, are unremarkable, save perhaps for the absence of all the other letters of the alphabet. Yet the specific sequence of these four letters represents an organism's genetic code, or genome, which underlies physical features and functions.
Making connections between the structures and functions of the genes encoded in the genome sequence is part of the integrative science mission of the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab). And one of the tools that researchers can apply to study the transcription factors that control how genes are turned on is known as DNA affinity purification sequencing or DAP-seq. The technology was developed by Ronan O'Malley, who leads the Sequencing Technologies Group. In an article published November 25, 2021 in Nature Methods, JGI researchers led by co-first authors Leo Baumgart and Juna Lee developed two approaches that build upon the DAP-seq technology O'Malley developed.
Unique capability
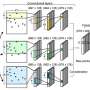
DAP-seq allows researchers to identify all the sites where transcription factors bind quickly and efficiently in the genome by adding tagged transcription factors to a genomic DNA library. "It's an innovative way to rapidly capture the binding location for most transcription factors in a species," O'Malley said. "It's a unique capability; no one else can do it in the world at this scale."
A bottleneck in the DAP-seq protocol, however, is the need to purify each transcription factor of interest. Nature Methods paper coauthor Lee did the initial development of a streamlined workflow that reduces the time and cost associated with this purification step, and Baumgart continued and expanded on the work known as biotin DAP-seq after she left the JGI.
"Biotin DAP-seq is a unique quick protein purification approach," O'Malley said. "It provides the transcription factors that you're going to need to then probe the genomic DNA for the binding sites." The technology expresses the transcription factor proteins from a DNA template amplified directly from either genomic DNA or complementary DNA (cDNA), reducing the amount of time it takes to produce a dataset from months to days and halving the total reagent cost.
While Biotin DAP-seq can be used by itself, it can also serve as a stepping stone to studying many genomes simultaneously through multiDAP, which allows researchers to conduct comparative analyses across the genomes of multiple species in a single experiment. "Effectively, you can identify conserved binding patterns shared between genomes of different species. This can provide insights into how they orchestrate sets of genes to execute similar functions. It can also identify cases where transcription factors have been repurposed during evolution to control new functions," O'Malley said. "Combining the two approaches of Biotin DAP-seq and multiDAP allows for ultra high throughput discovery of transcription factor binding sites across many different genomes. These atlases of transcription factor binding sites can help you better understand known biological functions as well as discover new functions. To help drive JGI User science we have implemented these two new methods as a high-throughput JGI capability supported by liquid-handling robotics."
Helps drive user science
DAP-Seq is already being applied to a number of approved proposals, including one led by Laszlo Nagy, a principal investigator at the Szeged Biological Research Center in Hungary. His approved proposal through the JGI's Community Science Program (CSP) focuses on a fungal comparative ENCODE project known as FUNCODE. "We noticed a few years ago that there is no shortage of fungal genomes anymore, but their functional interpretation is as tough as it has been 10 years ago," he said. "We thought about reconstructing gene regulatory networks, understanding where transcription factors bind in the genome. A key aspect of the project is understanding what TFs and regulatory networks are conserved across fungi."
Using multiDAP-seq, Nagy's team is comparing five fungal species to answer questions about how they break down plant materials, which could be useful for industrial biofuel production, and multicellular development. "DAP-Seq is the main source of information in the project," Nagy added. "Since the FUNCODE is primarily interested in transcription factor binding, we build a lot on DAP results. We also employ RNA-Seq and diverse in silico approaches for reconstructing gene regulatory networks."To learn more about how researchers have used JGI capabilities to further their research, watch the JGI Engagement Webinars series. Letters of Intent for the next annual Community Science Program proposal call are due in Spring 2022.
More information: Leo A. Baumgart et al, Persistence and plasticity in bacterial gene regulation, Nature Methods (2021). DOI: 10.1038/s41592-021-01312-2
Journal information: Nature Methods
Provided by DOE/Joint Genome Institute