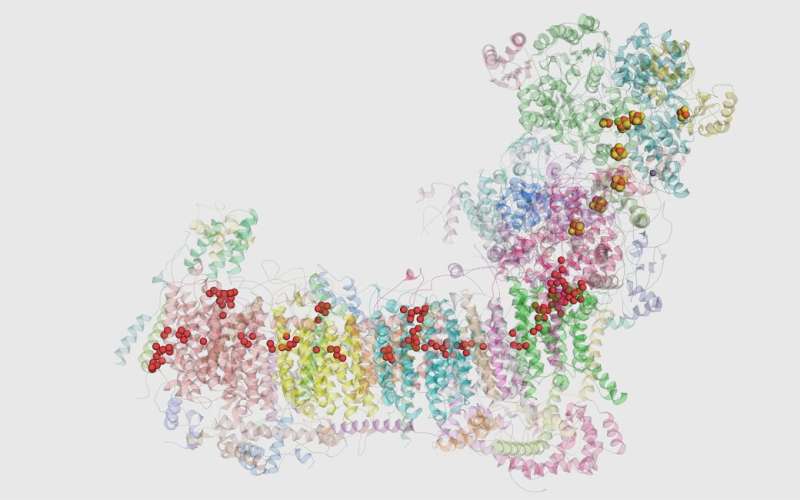
High-resolution dynamic insights obtained by computer simulations allowed researchers to identify molecular valves and novel design features in proteins. Credit: University of Helsinki
Structural biology and computer simulations pave the way for a deeper understanding of biological energy conversion. High-resolution structure and atomistic simulations allowed researchers to identify novel features in one of the key enzymes that contributes to energy generation in cells.
Energy (ATP) generation in the mitochondria of cells is one of the most important biological processes. The molecular understanding of this key metabolic pathway remains unclear, despite its central importance in biomedical and energy sciences. In a new study, researchers provide a detailed atomic view of one key component of mitochondrial energy metabolism—respiratory complex I.
The first enzyme in the electron transport chain of mitochondria, a pathway powering ATP synthesis, is respiratory complex I. This enzyme makes a significant contribution to ATP generation and is a focal point of mitochondrial function and dysfunction. Building upon their earlier joint collaborative studies (1,2,3), the researchers from the Goethe University (Frankfurt, Germany), the Max Planck Institute for Biophysics (Frankfurt, Germany) and the Department of Physics (University of Helsinki, Finland) have just provided the most detailed atomistic insight into the structure of complex I—a high resolution structure solved at 2.1 Å.
The high-resolution structure of a membrane protein of this size (~1 mega Dalton) is a remarkable feat in the field of membrane structural biology and mitochondrial biology. Researchers from Goethe University and the Max Planck Institute for Biophysics applied state-of-the-art cryo-electron microscopy to resolve the positions of several protein-bound water molecules, which carry out necessary proton transfer reactions required to convert chemical energy into ATP.
Jonathan Lasham, doctoral student from the Multi-scale modeling and Simulation of Mitochondrial Proteins research group at the University of Helsinki, performed large-scale molecular dynamics simulations of the high-resolution structural data. These simulations revealed a tight coupling between protein hydration, conformational dynamics and charge state of the protein. Currently, such high-resolution dynamic insights can only be obtained by computer simulations; this allowed researchers to identify molecular valves and novel design features in proteins, which are important in achieving high catalytic efficiency. Additional calculations and analysis by research assistant Amina Djurabekova and doctoral student Outi Haapanen from the same research group further consolidated the research results.
"This is yet another major step forward in our on-going collaboration with colleagues from Germany and in our understanding of biological energy generation, in that how respiratory complex I—a biomedically relevant enzyme—generates energy in the most efficient way," says Vivek Sharma, Sigrid Jusélius Senior Researcher from the Department of Physics at the University of Helsinki and PI of the Multi-scale Modeling and Simulation of Mitochondrial Proteins research group.
More information: Kristian Parey et al, High-resolution structure and dynamics of mitochondrial complex I—Insights into the proton pumping mechanism, Science Advances (2021). DOI: 10.1126/sciadv.abj3221
Etienne Galemou Yoga et al, Mutations in a conserved loop in the PSST subunit of respiratory complex I affect ubiquinone binding and dynamics, Biochimica et Biophysica Acta (BBA) - Bioenergetics (2019). DOI: 10.1016/j.bbabio.2019.06.006
Kristian Parey et al, High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease, Science Advances (2019). DOI: 10.1126/sciadv.aax9484
Etienne Galemou Yoga et al, Essential role of accessory subunit LYRM6 in the mechanism of mitochondrial complex I, Nature Communications (2020). DOI: 10.1038/s41467-020-19778-7
Journal information: Science Advances , Nature Communications
Provided by University of Helsinki