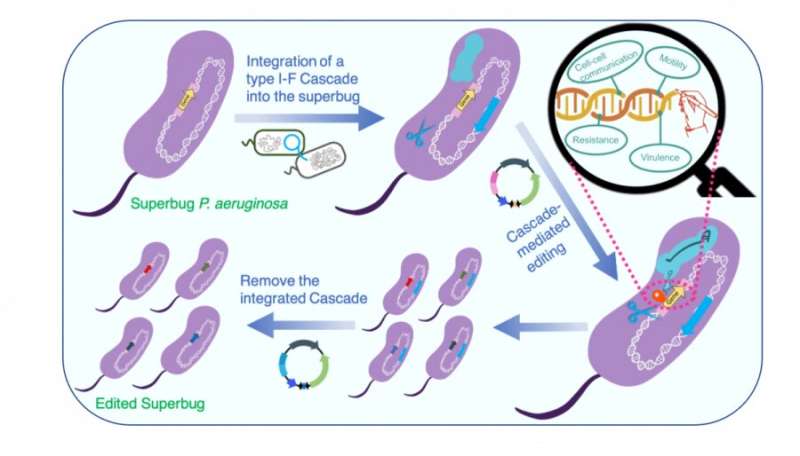
Strategies and workflow of the transferrable type I CRISPR Cascade-mediated editing of the superbug P. aeruginosa. Credit: The University of Hong Kong
A research team led by Dr. Aixin Yan, Associate Professor from the Research Division for Molecular & Cell Biology, Faculty of Science, in collaboration with Honorary Clinical Professor Patrick Cy Woo from the Department of Microbiology, Li Ka Shing Faculty of Medicine, the University of Hong Kong (HKU), reported the development of a transferrable and integrative type I CRISPR-based platform that can efficiently edit the diverse clinical isolates ofPseudomonas aeruginosa, a superbug capable of infecting various tissues and organs and a major source of nosocomial infections. The technique can accelerate the identification of resistance determinants of multidrug resistant (MDR) pathogens and the development of novel anti-resistance strategies.
The research opened a new avenue to genomically edit those wild bacterial species and isolates, such as those with clinical and environmental significance and those forming human microbiome. It also provided a framework to harness other CRISPR-Cas systems widespread in prokaryotic genomes and expand the CRISPR-based toolkits. The research has been published in theleading science journal Nucleic Acids Research.
Background
CRISPR-Cas system comprises the adaptive immune system in prokaryotes that disarms invading viruses by cleaving their DNA. Owing to its unique capability of targeting and altering DNA sequences, CRISPR-Cas has been exploited as the next-generation genome editing method. The method is based on the Class 2 type II CRISPR/Cas9 system, which has revolutionized genetics and biomedical research in a plethora of organisms and was awarded the 2020 Nobel Prize in Chemistry. However, the Class 2 CRISPR-Cas systems represent only ∼10% of the CRISPR-Cas systems encoded naturally in prokaryotes. Their applications to edit bacterial genomes are rather limited.
Remarkably, CRISPR-Cas systems belonging to different classes and types are continuously identified, and they serve as a deep reservoir for the expansion of the CRISPR-based toolkits. The most diverse and widely distributed CRISPR-Cas systems is the type I system which accounts for 50% of all CRISPR-Cas systems identified and has the potential to expand the CRISPR-based toolkits with distinctive advantages not accessible with the class 2 systems, such as high specificity, minimal off-targeting, and capable of large fragment deletions. However, type I CRISPR-Cas system hinges on a multi-component effector complex termed as Cascade to interfere DNA which is not readily transferrable to heterologous hosts, hindering the widespread application of these naturally abundant CRISPR for genome editing and therapeutics.
Key findings
Previously, the team has identified a highly active type I-F CRISPR-Cas system in a clinical multidrug resistantP. aeruginosastrain PA154197 which was isolated from a bloodstream infection case in Queen Mary Hospital. They characterized this CRISPR-Cas system and successfully developed a genome-editing method applicable in the MDR isolate based on this native type I-F CRISPR-Cas system. The method enabled rapid identification of the resistance determinants of the MDR clinical isolate and the development of a novel anti-resistance strategy (Cell Reports, 2019, 29, 1707-1717).
To overcome the barrier of transferring the complex type I Cascade to heterologous hosts, in this study, the team cloned the entire type I-Fcasoperon into an integration proficient vector mini-CTX and delivered the cassette into heterologous hosts by conjugation, a DNA transfer approach common in nature. The mini-CTX vector enabled the integration of the entire Cascade onto the conservedattBgenetic locus in the genome of the heterologous hosts, enabling them to harbor a "native" type I-F CRISPR-Cas system that can be stably expressed and function. The team showed that the transferred type I-F Cascade displays a significantly greater DNA interference capacity and higher strain stability than the transferrable Cas9 system and can be employed for genome editing with efficiency (>80%) and simplicity, i.e. by a one-step transformation of a single editing plasmid.
Furthermore, they developed an advanced transferrable system that includes both a highly active type I-F Cascade and a recombinase to promote the application of the system in strains with a poor homologous recombination capacity, wildP. aeruginosaisolates without genome sequence information, and in otherPseudomonasspecies. Lastly, the introduced type I-F Cascade genes can be readily removed from the host genomes through the I-F Cascade-mediated deletion of large DNA fragments, resulting in scarless genome editing in the host cells. The application of the transferrable system for gene repression was also demonstrated, highlighting the robust and diverse applications of the developed transferrable type I-F CRISPR system.
Dr. Aixin Yan predicted that this novel method will be extended to editing not only pathogens but also microbiome to promote human health, she said: "We believe that CRISPR-based technology and therapies will bring new hopes tocombattingsuperbugs in the future."
More information: Zeling Xu et al, A transferrable and integrative type I-F Cascade for heterologous genome editing and transcription modulation, Nucleic Acids Research (2021). DOI: 10.1093/nar/gkab521
Journal information: Nucleic Acids Research , Cell Reports
Provided by The University of Hong Kong