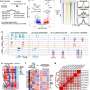
Feng Yue, PhD, the Duane and Susan Burnham Professor of Molecular Medicine, associate professor of Biochemistry and Molecular Genetics and of Pathology, director of the Center for Cancer Genomics at the Lurie Cancer Center, and director of the Center for Advanced Molecular Analysis in the Institute for Augmented Intelligence in Medicine, was senior author of the study published in Nature Methods. Credit: Northwestern University
Northwestern Medicine scientists have invented a new method for resolving rearranged chromosomes and their 3D structures in cancer cells, which can reveal key gene regulators that lead to the development of tumors, published in Nature Methods.
Feng Yue, Ph.D., the Duane and Susan Burnham Professor of Molecular Medicine and senior author of the study, said this method could help identify new targets for therapy.
"Cancer genomes contain tons of rearrangement. We frequently observe that big chunk of DNA fragments are lost, duplicated, or shuffled to another chromosome. Many of the known oncogenes and tumor suppressors are affected by such events," said Yue, who is also an associate professor of Biochemistry and Molecular Genetics, of Pathology and the director of the Center for Cancer Genomics at the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
Within each cell, strands of DNA, which if laid flat would stretch over two meters long, need to be properly folded and organized so that they can fit inside the nucleus, which is usually only a few micrometers in diameter. Often, DNA ends up forming "loops" that bring together genomic elements that are usually very far apart when the entire genome is unfurled.
Normal human cells have 46 chromosomes, but cancerous cells often have more, with many chromosomes cut into pieces and fused with parts from other chromosomes. Sometimes a cancer-specific chromosome is formed by stitching pieces of DNA from several different chromosomes, according to Yue.
"It is extremely challenging to figure out the composition of cancer genomes," said Yue, who is also the director of the Center for Advanced Molecular Analysis at the Institute for Augmented Intelligence in Medicine.
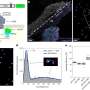
Fusion events, in combination with chromatin loops within these cancer-specific chromosomes, can lead to activation of oncogenes, but until now there was no systematic way to find the link between the two. To remedy this, Yue and his group members developed "NeoLoopFinder," a complete computational framework that analyzes the 3D structure of cancer chromosomes and identifies the essential regulators for key oncogenes.
It does so by resolving the complex genome rearrangement events, reconstructing the local chromatin interaction maps, estimating how many copies of such events in the cancer cells and finally by using a machine-learning algorithm they previously developed to find the chromatin loop.
"This helps us identify the control element for the oncogenes—the switch that turns the oncogene on or off," Yue said.
The importance of these loops in cancer has become increasingly recognized by the field, Yue said, with such events observed in nearly every type of cancer they analyzed. In the study, Northwestern Medicine scientists studied 50 different types of cancer cells, such as leukemia, breast cancer and prostate cancer. In two recently published studies, Yue and collaborators also used this method to examine chromatin loops in pediatric brain cancer and bladder cancer.
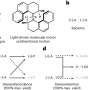
Using NeoLoopFinder, the investigators compiled a database of control elements—mainly enhancers—associated with oncogenes. As a proof-of-concept, they used CRISPR-Cas9 genome editing to disable a gene enhancer linked to an oncogene (ETV1) for prostate cancer, and found that disabling the enhancer shut down the oncogene.
"Normally you wouldn't imagine that disabling an enhancer on chromosome 14 would shut down an oncogene on chromosome 7, but now with our tool, more scientists will be able to identify such events and dissect their clinical implications in cancer," Yue said.
The computational framework is freely available to researchers at GitHub.
More information: Xiaotao Wang et al, Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes, Nature Methods (2021). DOI: 10.1038/s41592-021-01164-w
Journal information: Nature Methods
Provided by Northwestern University