Antibody development against medically important target proteins
Perfectly controlled protein scissors at the molecular level reveal the binding sites of antibodies, and enable the bottom-up design of new monoclonal antibodies aimed at medically important target proteins.
Karolinska Institutet, the Swedish biotechnology company Oblique Therapeutics AB, and Göteborg University, have, on April 16, 2021, published research results in the multidisciplinary scientific journal Science Advances, under the heading " Rational Antibody design for Undruggable Targets using Kinetically Controlled Biomolecular Probes."
This peer-reviewed article describes how Oblique Therapeutics' AbiProt technology, based on nanotechnology and proteomics, can be used to produce and develop pharmacologically tailored antibodies, directed to clinically important target proteins related to diseases that are generally considered impossible to treat with antibodies.
The results may be of great importance for Swedish research in biological pharmaceuticals, and Swedish drug development, as well as for millions of patients who are in need of new or improved medicines.
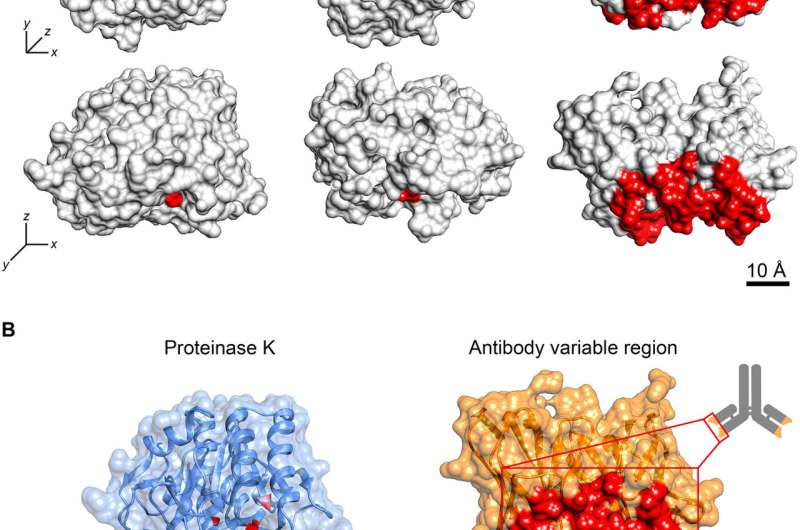
"We have developed a completely new approach for producing antibodies, by using an unique combination of high-end technology developed within this project, algorithms, and biological mass spectroscopy (proteomics). We have turned traditional antibody development upside down and asked ourselves—can one begin the development of an antibody by first understanding or localizing its binding site on a protein? The breakthrough is that we have direct access to high-resolution molecular information in the form of sequenced binding sites for antibodies (compare; sequencing in DNA and RNA). To obtain this information, we use protein scissors or proteases to probe antibody binding sites on a protein based on size similarity. Then we can use this sequence information for scalable drug development (compare; for example, recombinant DNA technology and CRISP-CAS9)," says Group Leader at Department of Physiology and Pharmacology, KI and CEO of Oblique Therapeutics, Professor Owe Orwar, who has manage the study.
"An important aspect of the invention is that the technology can be applied directly to native drug targets. We can therefore work with disease-relevant proteins that are in dynamic structures and forms, difficult to capture with other technologies. The high-resolution information we obtain about the nature of the binding site at the molecular level (i.e. individual amino acids in peptide sequences, so-called epitopes on proteins) means that in most cases we can identify a high-affinity antibody binding to a specific epitope," says Dr. Carolina Trkulja, first author and CSO, Oblique Therapeutics.
"From this detailed information, at the molecular level, we can then design antigens that are used for immunization or screening (e.g. phage display). The pharmacological profile of the antibodies can in some cases be programmed from the beginning and we can produce, for example, antagonistic or agonistic antibodies to many target proteins. This is a large medical/pharmacological breakthrough, and unlike traditional antibody techniques that are applicable to a few pharmacological targets or target proteins—AbiProt can address virtually the entire drug-resistant human proteome ", says Associate Professor Kent Jardemark, Group Leader at Deptartment of Physiology and Pharmacology.
Two examples of such antibodies are presented in the article. One type of antibody is against hTRPV1, a clinically validated target protein for the treatment of pain. The second type is targeting KRAS, a highly relevant oncogenic protein critical in the etiology of many aggressive cancers (e.g. pancreatic cancer). This target protein have previously been considered "undruggable" with antibodies. These early findings have the potential to contribute to the development of new antibody-based drugs for several treatment areas with great medical needs.
"KRAS is mutated in 80% of pancreatic cancers and 30% of colorectal cancer cases. This research means that we are one step closer to developing a more effective pharmacological treatment to combat these large groups of KRAS-mutated cancers," says Dr. Monica Marcus, Deptartment of Physiology and Pharmacology.
"We hope that the work in the long term will lead to a number of new antibody therapies against large disease areas that today lack effective therapies" says first author Oscar Jungholm, Ph.D. student at Deptartment of Physiology and Pharmacology.
Other antibodies in development include: TRPA1 (pain, cough), Trx (metastatic cancer) and SARS-CoV2.
More information: Carolina L. Trkulja et al. Rational antibody design for undruggable targets using kinetically controlled biomolecular probes, Science Advances (2021). DOI: 10.1126/sciadv.abe6397
Journal information: Science Advances
Provided by Karolinska Institutet