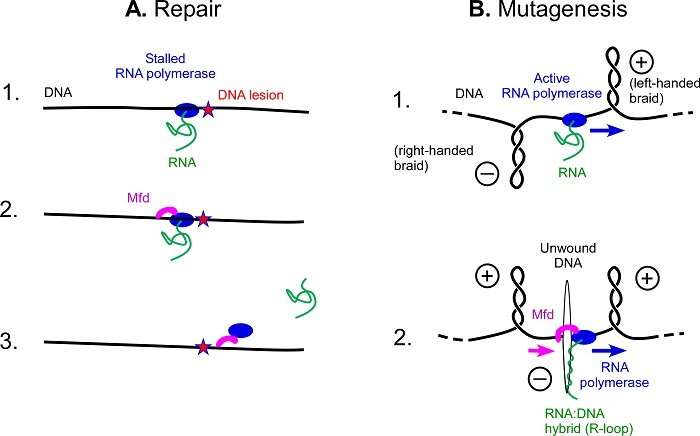
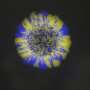
Schematic view of repair or mutagenesis processes. (A) Mfd repair function. 1. An RNA polymerase is blocked by UV damage in DNA. 2. Mfd binds to RNA polymerase and DNA, and moves along the DNA to remove the RNA polymerase from the DNA. 3. By removing RNA polymerase, RNA is released and Mfd continues on its way, recruiting more DNA repair factors. (B) Mfd mutagenesis function. 1. When an RNA polymerase travels through the DNA to read its inscriptions, it causes the accumulation of left-handed braids (corresponding to overtwisted DNA) ahead of the protein and right-handed braids (corresponding to undertwisted DNA behind the protein. 2. When Mfd interacts with an active RNA polymerase and binds to DNA, the right-handed braids are concentrated in the small area between Mfd and the RNA polymerase, causing the DNA to unwind. RNA can then be inserted into the double helix and form a DNA:RNA hybrid, the R-loop. Credit: Terence Strick
Using a specialized protein, all bacteria are capable of rapidly and effectively repairing damage to their DNA from UV. However, this mutation frequency decline (Mfd) protein plays another role and causes mutations. A team1 involving scientists from CNRS, ENS-PSL and supported by Inserm has demonstrated and described this phenomenon. Better understanding of mutations opens up prospects in the fight against resistance to antibiotics and anti-cancer therapies. This study was published in PNAS on 5 April 2021.
DNA can undergo many degradations, including UV degradation. Just as a landslide on a railway track will prevent a train from passing, the damage generated by UV rays is an obstacle to RNA polymerase, a protein that travels along the length of DNA to read its instructions. RNA polymerase being blocked at any point prevents damage from being repaired. To restore the "track", bacteria possess the Mfd protein that clears the blocked RNA polymerase, and then recruits other proteins to help with the repairs, avoiding mutations.
Biologists from the Institut de Biologie de l'ENS (CNRS/ENS—PSL/INSERM) have just solved the mystery behind a previously lesser-known role of this protein. It is involved in the formation of DNA areas where mutations appear more easily.
The cell machinery involving the Mfd protein normally repairs DNA, but when a bacterium faces certain stressors, such as the presence of an antibiotic, it takes another route: Mfd grips to an active RNA polymerase as it scrolls through DNA, rather than clearing a blocked RNA polymerase. Once bound together, the two proteins unwind the double DNA helix rapidly and on a large scale, forcing it open. The opening of the two DNA strands creates a favorable environment for the formation of particular zones facilitating mutations: R-loops.
R-loops are still poorly understood structures, but they are the source of many mutations: scientists have shown that nearly half of the mutations in bacteria are due to these structures. As antibiotic resistance is born from mutations, this work, which demonstrates the link between Mfd and R-loops, opens up new perspectives in the fight against antibiotic resistance.
The research team also points out that Mfd has equivalents in the various branches of life and that this mutation mechanism is probably universally present. In humans, for example, the protein homologous to Mfd is involved in accelerated aging and evolution of tumor resistance to chemotherapy. Scientists believe that a mechanism similar to R-loop formation could occur in cancer cells, introducing mutations that can make them resistant to treatment.
More information: Co-transcriptional R-loop formation by Mfd involves topological partitioning of DNA. James R. Portman, Gwendolyn M. Brouwer, Jack Bollins, Nigel J. Savery, and Terence R. Strick. PNAS, April 5, 2021. DOI: 10.1073/pnas.2019630118
Journal information: Proceedings of the National Academy of Sciences
Provided by CNRS