'Designer' pore shows selective traffic to and from the cell nucleus
The nucleus is the headquarters of a cell and molecules constantly move across the nuclear membrane through pores. The transport of these molecules is both selective and fast; some 1,000 molecules per second can move in or out. Scientists from the University of Groningen and Delft University of Technology, both in the Netherlands, and a colleague from the Swedish Chalmers University of Technology, have developed an artificial model of these pores using simple design rules, which enabled them to study how this feat is accomplished. Their results were published on 31 March in Nature Communications.
Nuclear pores are extremely complicated structures. The pore itself is a big protein complex and the opening of the pore is filled with a dense network of disordered proteins called nucleoporins. These proteins regulate selective transport, but exactly how they do this is still unclear. "The nuclear pore complex is one of the biggest protein structures in the cell," explains Patrick Onck, professor of Micromechanics at the University of Groningen. "We previously studied the pores in all their complexity, but for this study, we created a drastically simplified 'designer' pore to investigate the essential physical mechanisms of transport."
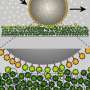
Nanopore
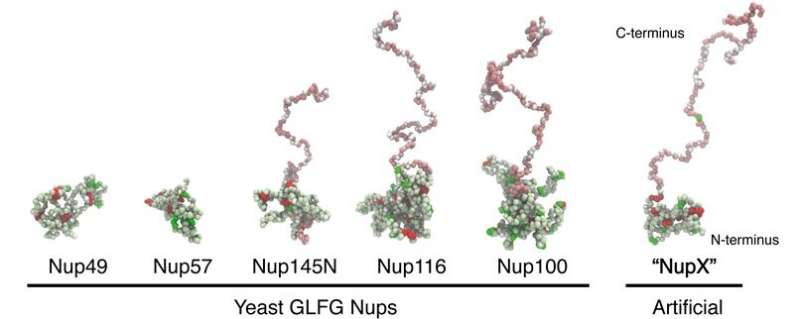
First, the team analyzed the composition of the nucleoporins to design a simplified, 'average' version, which they termed nucleoporin X, or NupX for short. These proteins are made up of domains comprising phenylalanine (F) and glycine (G) amino acids in tandem, and these play an essential role in transport. These FG repeats are separated by 'spacers' of other amino acids. In addition to the FG repeats, some nucleoporins also contain domains of glycine, leucine, phenylalanine and glycine, or GLFG repeats. The team designed proteins that contain both domains, separated by spacers of ten amino acids.
NupX was tested in two different systems: it was studied experimentally, attached to a surface and added to artificial nanopores that were 'drilled' in a 'membrane' of silicon nitride, and through molecular dynamics simulations. The experiments were performed at Delft University of Technology, while the simulations were prepared and executed in Groningen, mostly by Henry de Vries, a Ph.D. student in Onck's laboratory.
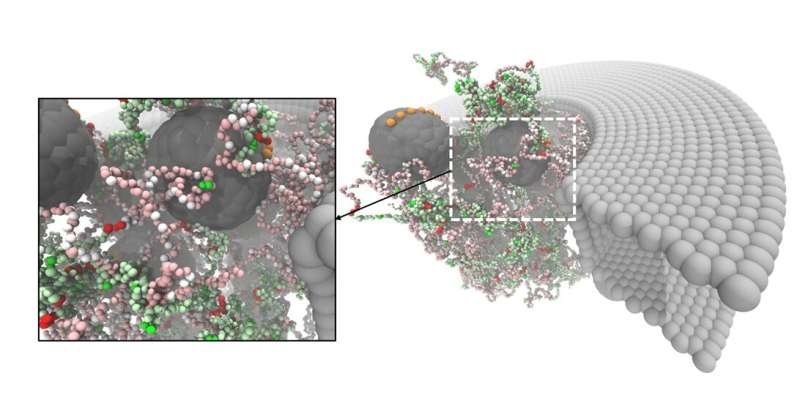
Transport ticket
The nucleoporins were tested for interactions with non-specific proteins and with chaperones, which are proteins that act as transport tickets through the pore. In the cell, large molecules that must be transported into or out of the nucleus can only do so when they are attached to such a chaperone. The artificial nucleoporins selectively interacted with the chaperones but not with the non-specific proteins. This demonstrated that the NupX pores are fully functional: they are able to facilitate selective transport. De Vries: "However, the experiments showed that transport through the artificial pores occurs but not what happens inside the pore. With our simulations, we showed what exactly happens inside the pore as the chaperones translocate, while the non-specific proteins do not interact with the pore at all."
The simulations also revealed how the FG and the GLFG nucleoporins were distributed inside the pore. "Recent studies suggested that they would be in different places in nuclear pores and that this might help to create selectivity," says De Vries. "However, we found that they were homogenously distributed and yet we still saw selectivity." Another suggestion was that the amino acids that make up the spacers are important for the selectivity. "Our results showed that the specific sequence of amino acids in the spacer doesn't matter since we used random sequences. The only important part is the ratio of charged amino acids to hydrophobic amino acids within the spacers, which determines the stickiness of the proteins."
Redundancy
The final conclusion of the study is that a very simple system in nucleoporins that has limited variation still produces a selective pore. "What is needed is a certain density of these FG nucleoporins," says Onck. "These form a barrier, which can only be breached by the chaperones." This begs the question of why the pores contain a very large number of different nucleoporins in nature. Onck: "We know that nature doesn't always come up with optimized solutions. However, their redundancy could very well have a function in natural pores."
The fact that the very simple artificial system already reproduces selective transport mechanisms means that the scientists now have an excellent tool to study the physical principles that regulate nuclear pore function. Onck: "This could lead to new fundamental insights but also to new applications, for example in creating filtration systems, or in the design of artificial cells."
More information: Alessio Fragasso et al, A designer FG-Nup that reconstitutes the selective transport barrier of the nuclear pore complex, Nature Communications (2021). DOI: 10.1038/s41467-021-22293-y
Journal information: Nature Communications
Provided by University of Groningen