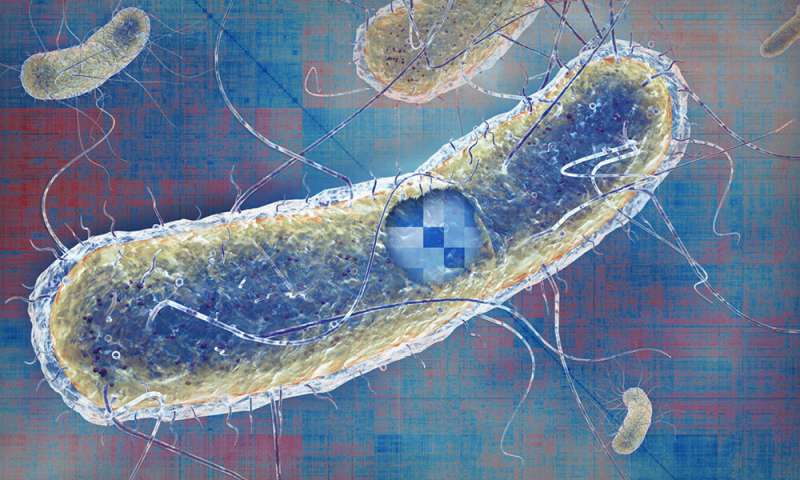
The temperature at which a protein denatures can give researchers clues about how it interacts with other molecules. Credit: Savitski team, Holly Joynes/EMBL
Understanding how genes work and how they interact with one another is a major goal of biology. This poses huge challenges in terms of both methods and the sheer numbers of experiments required. Recent advances have transformed scientists' ability to map gene function and interactions, and EMBL researchers are developing innovative techniques to measure the activity of thousands of genes at once. A new paper from EMBL's Savitski team and Typas group is published in Nature this week, describing their work on E. coli and how it brings a greater understanding of the way genes function and interact.
Thermal proteome profiling unravels the function of bacterial proteins
Central to this work is thermal proteome profiling (TPP), which was developed by EMBL team leader Mikhail Savitski in 2014. TPP is based on the principle that exposure to heat causes the structure of proteins to break down—they're said to 'denature' and they become insoluble. It's the same process that causes an egg white to become solid when cooked.
The team used the TPP approach on live E. coli bacteria, heating cells to different temperatures to investigate how much of each individual protein in the cell remained soluble at each temperature. The temperature at which a protein denatures can tell researchers about the state of a protein, whether it is active and/or interacting with, for example, a drug or another protein. This is because the structure of a protein changes when it takes part in any interaction, and different structures of the same protein require different levels of heat before they denature.
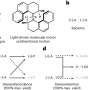
The team conducted this experiment on 121 E. coli mutants, with each having one gene removed. They profiled these mutants one by one using TPP, and looked at how all the proteins in the cell were expressed and the temperature at which they denatured. This information provided unique insights into the interactions and functions of each protein, including hundreds of less well-understood proteins.
The difficulty of working with essential proteins
One frequently used way to study individual proteins is to delete the genes encoding them and monitor the impact on how a cell behaves. Yet such an approach is impossible to use with essential proteins—those that a cell relies on for survival—because their absence would cause the cell to die. The technique used by the Typas and Savitski researchers in this study represents a big step forward in the study of essential proteins. "We found that although the levels of these proteins did not change across the mutants we studied—a feature which makes these proteins difficult to study by measuring their expression—their thermal stability changed quite often," says André Mateus, a postdoc in the Savitski team and Typas group, and first author of the paper. The change in thermal stability of essential proteins is because of changes in their activity.
While this research was conducted on E. coli—arguably the most studied bacterial species—the researchers hope to apply a similar approach to other organisms that are much less well understood. One such target is the human gut microbiome: the ecosystem made up of all the microorganisms in the gut. This ecosystem is a major focus of EMBL's future research plans, and is increasingly being linked to multiple aspects of human health and disease—everything from brain function to how our immune system operates.
More information: André Mateus et al. The functional proteome landscape of Escherichia coli, Nature (2020). DOI: 10.1038/s41586-020-3002-5
Journal information: Nature
Provided by European Molecular Biology Laboratory