Properly developed body plan of mouse embryos ensured by lysine demethylase 7a enzyme
The body plan of an organism, crafted over millennia of evolutionary trial and error, is so exquisitely fine-tuned that even a subtle deviation can be detrimental to individual survival and reproductive success. Now, researchers at the University of Tsukuba have elucidated the workings of an enzyme, lysine demethylase 7a (kdm7a), that facilitates appropriate development of the mouse embryo from tip to tail, 'according to plan' by modulating expression of Hox genes. As these genes, master regulators of embryonal morphogenesis, have been highly conserved over evolution, the findings apply in varying degree to lower species and to all vertebrates, us included.
It is an astounding fact that the unicellular zygote formed at fertilization contains all the information needed for development into a multicellular organism of immense complexity organized in well-ordered symmetry. How these data are encrypted and decoded is an escalating mystery as emerging answers only unearth further questions. Hox genes allocate regions along the head-tail axis of the developing embryo for development of appropriate structures; in vertebrates they specify the numbers and sequential shapes of the spinal bones.
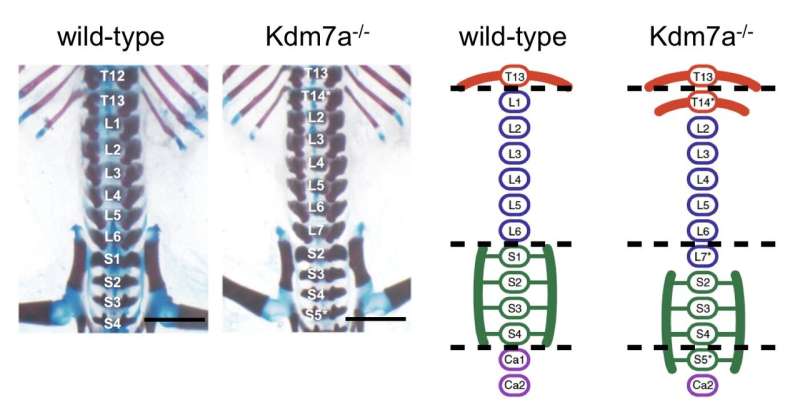
Some histone-modifying enzymes have been implicated in normal morphogenesis as well as in disease. Using CRISPR-Cas9 gene editing technology, the research team first developed knockout mice (Kdm7a−/−) by introducing frameshift mutation. As a result, they obtained mice carrying the mutations for truncated Kdm7a proteins lacking demethylase activity.
The researchers analyzed postnatal skeletal preparations of both wild-type and Kdm7a-/-mice. Dr. Yasuharu Kanki, senior author, describes the findings. "As expected, all wild-type mice showed a normal axial skeleton. Interestingly, all Kdm7a−/−mice and some heterozygous mutants exhibited vertebral transformation; some vertebrae assumed the characteristics and appendages of their anterior neighbors."
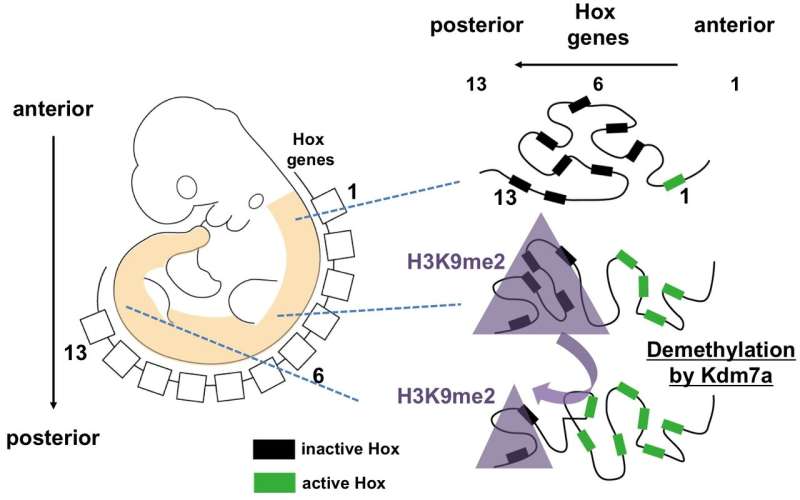
The researchers next used RNA sequencing to examine the expression of Hox genes during embryogenesis. Their findings support a functional role of Kdm7a-mediated transcriptional control, especially of the posteriorly situated Hox genes, and suggest that regulation of the repressive histone mark H3K9me2 might be involved.
"Our data help explain morphogenesis along the anterior/posterior axis in the mouse embryo and, by extension, in all vertebrates including humans," says Dr. Kanki. "Deciphering the interplay of various genetic and epigenetic determinants of embryonal morphogenesis as well as the underlying molecular mechanisms increases our knowledge of evolutionary developmental biology and may help in the understanding of disease."
More information: Yoshiki Higashijima et al. Lysine demethylase 7a regulates murine anterior-posterior development by modulating the transcription of Hox gene cluster, Communications Biology (2020). DOI: 10.1038/s42003-020-01456-5
Journal information: Communications Biology
Provided by University of Tsukuba