December 7, 2020 report
Introducing MitoCarta 3.0, the definitive guide to the elusive mitochondrial proteome
One of the most hotly contested issues in biology, or at least in mitochondrial biology, is determining which proteins can get into mitochondria. Having this kind of access essentially means a protein maintains a residence in one or more of several distinct mitochondrial compartments. These places include the matrix, the inner membrane, the intermembrane space, the outer membrane and the most nebulous listing of them all—an association with the outer membrane. Typically, it is this location that draws the biggest controversy.
The current industry standard mitochondrial inventory is MitoCarto, which is maintained at Harvard and MIT's Broad Institute. It currently lists 1,136 mitochondrial proteins distributed across 14 tissue types. Over the years, its ranks have swelled as new members were welcomed with each successive version. However, with the latest release of MitoCarta 3.0, 100 questionable protein coding genes were kicked out, while only 78 new ones were added. The other major upgrade is the addition of annotations for the sub-mitochondrial localization, and the assignment of genes to a newly emerged concept called gene ontology, which here includes 149 metabolic pathways.
Most ontologies and sub-ontologies (like fatty acid synthesis, complex IV, mtRNA metabolism or Fe-S cluster synthesis) are fairly well-defined and incontrovertible. But many are still unavoidably redundant, with bifunctional proteins necessarily assigned to multiple ontologies. One such ontology, the whimsically named "mitochondrial central dogma," remains vague, as it includes everything from mt-rRNA modifications and ribosome assembly to translation factors. Maintaining rigor here is no trifling matter as countless labs around the world rely on this database in the race to decode life itself, and by implication, rid the world of disease.
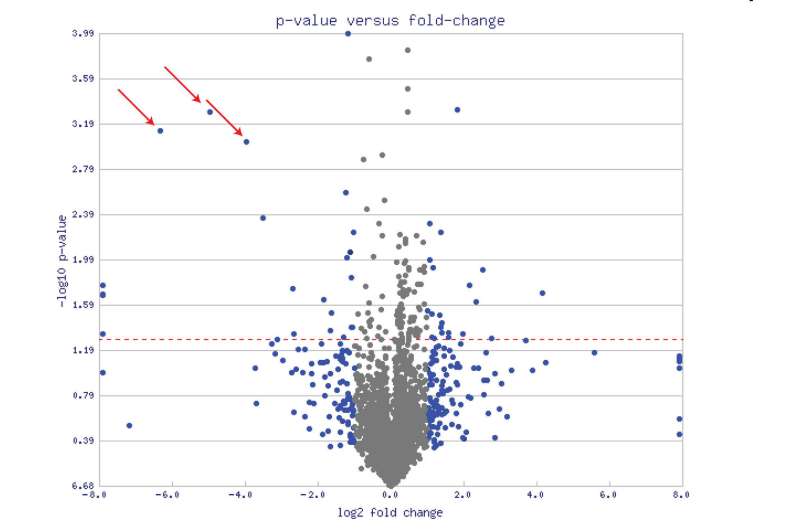
But we are not here to nitpick. Let's take MitoCarta 3.0 out for a test drive and get into some real-world examples. One paper that has brought the full power of MitoCata to bear was recently published in the journal Cell Stem Cell. Deploying the list from MitoCarta (2.0), the researchers used a technique called mass spectrometry proteomics to identify differences between neurons and astrocytes. Typically, proteomics results are presented in the form of a volcano plot (see the main picture above for an example). The fatter the volcano plume, the more protein levels either increased and/or decreased.
In the end, the researchers found around 150 mitochondrial proteins that were uniquely enriched in either cell type. They were then able to CRISPR engineer the most significant of the neuron-specific proteins (like for example, SOD-1) into astrocytes, and miraculously reprogram these astrocytes into neurons. One of the limitations of MitoCarta, and for that matter, almost every other "Ohmics" database out there is a lack of precision in declaring cell types or locations in the nervous system. This is due to the fact that neurons of almost every transmitter variety can be found in almost every large, distinctive structure in the brain. While some databases do include loose categorizations for inhibitory interneurons or perhaps excitatory cortical neurons, MitoCarta users must still make due with the rather broad cerebrum/cerebellum/brainstem/spinal cord classifications.
In this void, what is really needed is for someone to make a dedicated NeuroCarta1.0 to fill in the missing cell-type data for the entire nervous system. Also, since it is in the season, a searchable database to check for the presence of specific proteins rather than having to scan through all of them manually would make a nice gift. The proteomics paper mentioned above is a great first start in this general direction.
Last Thursday, we published a review of mitochondrial NAD metabolism, with a specific focus on the molecule CD38 . CD38 has emerged as a premier molecule for controlling mitochondrial transfer between astrocytes, and studies have claimed that it localizes to mitochondria. The same claims have been made for another molecule with a similar function called SARM1 in neurons. When I checked MitoCarta, I could not locate these proteins and assumed that they did not meet the stringent criteria for inclusion. I spoke with Vaamsi Mootha and Sara Calvo from the Broad Institute and they confirmed that these molecules are not included, but they assured me that the relevant papers will be reviewed in more detail during the next round of manual curation.
If I had to wager, I might guess there are perhaps somewhere between 1,500 and 2,000 molecules for which varying degrees of evidence for mitochondrial localization have been presented in the literature. Inclusion for these kinds of molecules really comes down to a manual inspection and replication of the data using other methods. Mootha and Calvo note that the 14 tissue types may have to be expanded to avoid missing certain proteins with highly specific expression. For example, IRG1, which encodes a mitochondrial aconitate decarboxylase that produces the immunometabolite itaconate is expressed almost exclusively in suitably activated macrophages.
I want to make a brief point here about the current state of affairs of peer review, a process that is integral to the present endeavor. I had reached out to contributing authors of the proteomics paper, Magdalena Gotz and Stefanie Hauck, because I simply could not make heads or tails of their supplemental Table 1 which purported to contain the actual protein level values for the astrocytes and neurons. They seemed to me to be all over the place, and without clear differences. I was relieved when they eventually responded to my emails and told me that upon looking again, they found the table was corrupted by a conversion incompatibility during the preparation of the English/German Excel data that had been obtained from calculation in the programming language R in the requisite .csv files. Okay, fair enough, but how did the actual reviewers not catch this while reviewing the paper?
Normally, I wouldn't have any concern about these kinds of readily explained errors everyone makes, and for the record, believe the above error has been fully explained. But curiously, the day before, I was reading a well-received paper published in the Quarterly Review of Biophysics that claims to have finally cracked the code, so to speak, for understanding the elusive origins of the mitochondrial amino acid codon table—the code of life itself, or at least of the major organelles driving eukaryogenesis. This is the signature evolutionary conundrum that has vexed researchers for decades. Again, I could not quite make sense of a figure that was illustrating a key point of the paper.
The issue was in Figure 2, which showed a hypercube representation of all possible single-step transitions and transversions for interconverting the entire codon table. Specifically, I had asked whether the upper right-hand vertex of the cube in the upper left-hand corner of the larger hypercube had a typo and should in fact be "AAA' instead of "GCA." Similarly to the case above, I was not immediately responded to when asking for clarification. Fortunately—and again, to the credit of the authors—with sufficient prodding, researcher Kenneth Breslauer eventually responded. He said thanks, and that an errata has been prepared to fix the oversight. He also gave assurance that when the actual analysis was done, the correct codon had been used. It looks like this error still stands in the publication for now, and will hopefully be amended soon.
But how is it possible that professional reviewers did not raise such basic, and frankly, essential questions that a woefully inexpert blogger has raised? Much to our regret, the answer must simply be that peer review as it now exists is not what we have been led to believe.
More information: Sneha Rath et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations, Nucleic Acids Research (2020). DOI: 10.1093/nar/gkaa1011
Journal information: Cell Stem Cell , Nucleic Acids Research
© 2020 Science X Network