Giving cells an appetite for viruses
A team led by UT Southwestern researchers has identified a key gene necessary for cells to consume and destroy viruses. The findings, reported online today in Nature, could lead to ways to manipulate this process to improve the immune system's ability to combat viral infections, such as those fueling the ongoing COVID-19 pandemic.
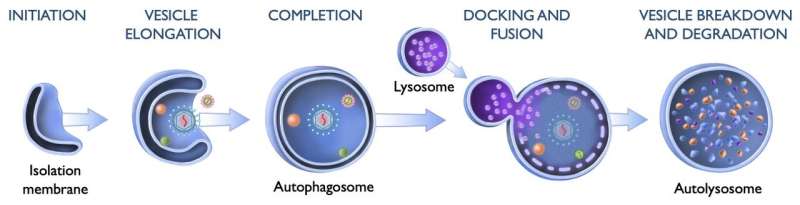
Scientists have long known that cells use a process called autophagy to rid themselves of unwanted material. Autophagy, which translates as "self-eating," involves isolating cellular garbage in double-layered vesicles called autophagosomes, which are then fused with single-layered vesicles known as lysosomes to degrade the materials inside and recycle them into building blocks for other uses.
This process helps cells discard old or defective organelles and protein complexes, bacteria, and viral invaders. For a variety of cellular refuse tasks, researchers have identified clear pathways by which cells initiate and regulate autophagy. But it's been unclear to date, notes study leader Xiaonan Dong, Ph.D., lead author of the study and assistant professor of internal medicine at UTSW, whether a unique pathway specifically targeted viruses for autophagy.
In this latest research, Dong and his colleagues manipulated human cells infected by different viruses to individually deplete more than 18,000 different genes, examining their effects on autophagy. As their initial models, the researchers infected cells with herpes simplex virus type 1 (HSV-1), which causes cold sores and sexually transmitted infections, and Sindbis virus, which causes a mosquito-borne illness most common in parts of Africa, Egypt, the Middle East, the Philippines, and Australia.
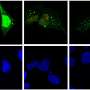
The team's investigation identified 216 genes that appear to play a role in viral autophagy. To narrow their search to the key players, the researchers used bioinformatics to analyze biological processes these genes regulate. They quickly honed in on a gene called sorting nexin 5 (SNX5), which produces a protein that helps recycle plasma membrane-anchored proteins from endosomes, sorting organelles in cells that often ferry materials isolated outside cells into their interior. Because viruses often enter cells through this pathway, the team reasoned, SNX5 could be important for their autophagy.
When the scientists shut down SNX5 in human cells, the cells' ability to perform autophagy on HSV-1 and Sindbis viruses significantly decreased—however, their ability to activate autophagy as part of normal cellular cleaning or for bacteria removal stayed intact, suggesting that this gene is specifically used for viral autophagy. This effect persisted for several other viruses, including Zika, West Nile, chikungunya, poliovirus, Coxsackievirus B3, and influenza A, suggesting that SNX5 is part of a general mechanism for autophagy across a broad range of viruses.
Further experiments showed that deleting SNX5 greatly increased susceptibility to infection in both lab-grown cells and in adult and juvenile animals. However, when these cells were infected by viruses manipulated to suppress their ability to induce autophagy, they were largely spared.
Together, explains Dong, these results suggest that cells do have a unique pathway for viral autophagy that likely has SNX5 at the helm. This finding not only solves a long-standing mystery in basic biology, he adds, but it could eventually lead to new ways of fighting viral infections. Most current methods target viruses through their individual weaknesses—an approach that necessitates developing a unique strategy for every viral type.
"By learning how cells naturally take up and degrade viruses," Dong says, "we could discover ways to augment this process, creating a more general strategy for developing broad-spectrum antiviral therapeutics that combat an array of different viral infections."
Co-leaders of this research effort were the late Beth Levine, M.D., former director of the Center for Autophagy Research and professor of internal medicine and microbiology at UT Southwestern, and Ramnik J. Xavier, M.D., the Kurt Isselbacher Professor of Medicine at Harvard Medical School and a core member of the Broad Institute of MIT and Harvard. Levine, a world-renowned leader in the study of autophagy, died in June. She also was an investigator with the Howard Hughes Medical Institute and member of the National Academy of Sciences. At UT Southwestern, she held the Charles Sprague Distinguished Chair in Biomedical Science.
"This is a beautiful study that further cements the legacy of Dr. Levine and work from her lab members," says Julie Pfeiffer, Ph.D., professor of microbiology at UTSW and a contributor to the study who holds the Kern and Marnie Wildenthal President's Research Council Professorship in Medical Science.
More information: Dong, X., Yang, Y., Zou, Z. et al. Sorting nexin 5 mediates virus-induced autophagy and immunity. Nature (2020). doi.org/10.1038/s41586-020-03056-z
Journal information: Nature
Provided by UT Southwestern Medical Center