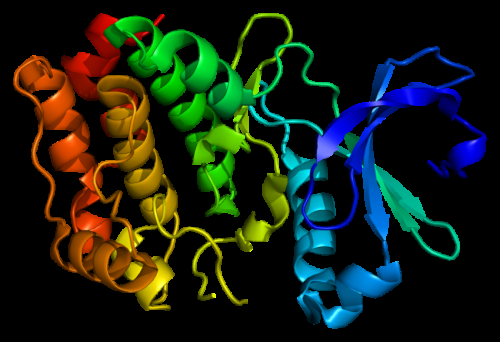
Structure of the Aurora A kinase protein. Based on PyMOL molecular visualization system rendering of Protein Data Bank (PDB) 1mq4. Credit: Created by Emw, Wikimedia CC BY-SA 3.0.
In order for tumors to successfully take hold and spread in the body, they typically must meet several developmental milestones. These include things like suppressing apoptosis, cultivating replication and angiogenesis, weaning from growth factor dependency, becoming less sticky, and becoming more sticky.
Behind each of these pillars of multicellular harmony, there is usually some kind of protein kinase pulling the strings. Often, these kinases turn rogue because of mutations that hyper- or hypo-activate them, or because of other nefarious events like gene fusions. Inhibiting the actions of these traitors has therefore become a mainstay of anti-cancer therapy.
Large online databases have emerged as essential tools in this endeavor. On Monday, we introduced and tested MitoCarta 3.0, a popular, freely available compendium of the entire mitochondrial proteome. While this proteome is still green with continuous additions and subtractions to its ranks, the big-data kinase world might be said to be a bit more mature.
Long-term support has been maintainedfor around 518 protein kinases since 2002 . This is essentially the human kinome. There are also around 55 partially defunct pseudokinase reserves still mothballed in genetic hangars. Many serve in scaffolding roles, while others persist largely in limbo, hoping one day that the potluck of genetic recombination might restore their former glory. The faults and features of each member of the kinome have been refined into special-purpose databases for tabulating mutations, inhibitors, interactions, structures and more. The latest major addition, whimsically dubbed KuNGFU for Kinase Gene Fusion Database, was just published in the journal Nature Scientific Data.
KuNGFU contains all the experimentally validated in-frame kinase gene fusions that retain active catalytic domains that have been identified in cancer cell lines. In-frame fusions preserve the codon structure of the fusion genes, and will therefore be translated into the proper amino acid sequences of the fusion proteins. These features offer a unique druggable set of kinase gene fusion targets in cancer models. Druggability is a nebulous term often used in a fashion similar to "drinkability," which of course, means a beer to which one can bind with high affinity—or at least one that appears to be similar to other previously characterized drinkable beers.
Databases like KuNG FU have been made possible by advances in deep-sequencing and fusion detection algorithms. One nice thing about it is that the freely available repository comes complete with open-source code. Users can add to the database as they see fit, and can also avail themselves of the Python scripts needed to set up and interact with their own MySQL database. The Github code is available here.
Another useful proteomics database known as the Global Map of Kinase Degradability has recently been published in the journal Cell. It is also open access, and provides chemical leads for over 200 kinases. Researchers have realized that the best way to squelch an offending kinase is not always to inhibit it with drugs. Since kinases all have mostly similar catalytic domains and structures, drugs will tend to have many side effects. A smarter way may be to degrade them naturally. In other words, to use a cell's existing proteosomal system (and its expansive suite of artisanal ubiquitin ligases) to selectively drive kinases onto the chopping block.
The is done using special small degrader molecules which are sometimes given the unfortunate name of PROTAC, for proteolysis targeting chimera. One end of the molecule, the warhead, binds to a specific ubiquitin E3 ligase. The other end, the target, binds to the undesired protein. If everything works out—that is to say, if the binding successfully targets, degrades, and establishes functional effect—then you've got a winner. This winner is not necessarily the guy with the highest binding affinity; instead, it is the guy that can do all of these steps. The up-to-date map of the degrading kinome can be taken out for spin over at the lab of Nathanael Gray and Eric Fischer at the Dana-Farber Cancer Institute.
Over at the biotech hotbed in Malvern, Pennsylvania, there are two companies with special expertise in these matters. One is Life Sensors, which is an industry leader in providing goods and services for studying ubiquitination and degraders. The other is Reaction Biology, which recently acquired ProQinase in Germany, together with their large panel of active recombinant protein kinases. Reaction Biology offers web tools of their own, including a Kinase Mapper for visualizing kinase activities.
The combination of omics resources into the promised intelligent multi-ohmic bliss awaits fruition. However, the rapid proliferation of openly accessible databases in corporate and academic biotech is certainly a bright spot on the year.
More information: Alessio Somaschini et al. Mining potentially actionable kinase gene fusions in cancer cell lines with the KuNG FU database, Scientific Data (2020). DOI: 10.1038/s41597-020-00761-2
Katherine A. Donovan et al. Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development, Cell (2020). DOI: 10.1016/j.cell.2020.10.038
Journal information: Cell
© 2020 Science X Network