Identification of a viral factor that impairs immune responses in COVID-19 patients
As of October 2020, SARS-CoV-2 is the cause of an ongoing pandemic, with more than 35 million reported cases and more than 1 million deaths worldwide. One prominent feature that distinguishes COVID-19 from SARS in terms of immune responses is the poor induction of a type I interferon (IFN) response by SARS-CoV-2 compared to SARS-CoV and influenza A virus. Notably, impaired IFN responses are associated with COVID-19 disease. However, the molecular mechanisms underlying the inefficient IFN responses in SARS-CoV-2 infection remain unclear.
A research team at The Institute of Medical Science, The University of Tokyo (IMSUT) aimed to characterize the viral factor(s) determining immune activation upon SARS-CoV-2 infection and found that ORF3b, a gene encoded by SARS-CoV-2, is a potent IFN antagonist.
"The poor IFN responses in COVID-19 patients may be explained by the action of this viral product, ORF3b", said the lead scientist, Kei Sato, Associate Professor (Principal Investigator) at Division of Systems Virology, Department of Infectious Disease Control, IMSUT.
The results of this research were published in Cell Reports on September 4, 2020.
ORF3b as a viral IFN antagonist
Although SARS-CoV infection causes acute and severe pneumonia, SARS-CoV-2 infection may be asymptomatic or result in flu-like symptoms such as fever, cough and fatigue. Also, compared to SARS-CoV and influenza A virus infections, a hallmark of SARS-CoV-2 infection, COVID-19, is the poor induction of a type I interferon (IFN). Notably, impaired IFN responses are associated with the severity of COVID-19. However, the molecular mechanisms underlying the inefficient IFN responses in SARS-CoV-2 infection remain unclear.
By comparing the sequences of SARS-CoV-2-encoding genes to those of SARS-CoV, the research group found that the gene length of SARS-CoV-2 ORF3b is markedly shorter than that of SARS-CoV ORF3b.
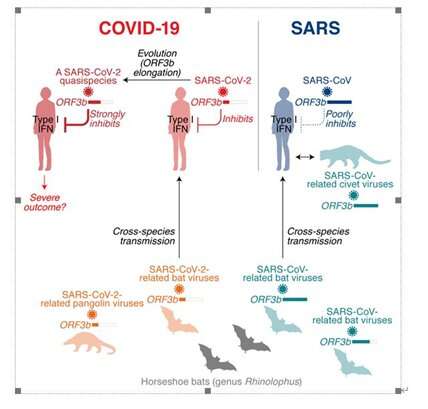
Because ORF3b of SARS-CoV is known as a viral antagonist against IFN production, they hypothesized that the difference on the length of ORF3b gene between SARS-CoV-2 and SARS-CoV may alter their anti-IFN activity and further may explain the difference in the symptoms of these two viral infections.
Surprisingly, SARS-CoV-2 ORF3b is a more potent IFN antagonist than SARS-CoV ORF3b. Phylogenetic analyses and functional assays revealed that SARS-CoV-2-related viruses from bats and pangolins also encode shorter ORF3b gene products with strong anti-IFN activity.
Characterization of a natural SARS-CoV-2 ORF3b variant with enhanced anti-IFN activity
Furthermore, analyses of approximately 17,000 SARS-CoV-2 sequences identified a natural variant, in which a longer ORF3b reading frame was reconstituted. This variant suppresses IFN even more efficiently than ORF3b of the parental SARS-CoV-2 strain.
In agreement with an association of IFN suppression with disease severity, the two patients in Ecuador harboring SARS-CoV-2 with the extended ORF3b variant were critically ill; one was treated in an intensive care unit and the other one died of COVID-19.
Importantly, however, there is no direct evidence indicating that the viruses detected in these two COVID-19 patients in Ecuador are more pathogenic than the reference strain. Although they cannot tell whether this variant is associated with a different outcome in disease, it is plausible that naturally occurring length variants of ORF3b can potentially contribute to the emergence of more pathogenic SARS-CoV-2 variants.
Thus, it will be important to continue monitoring viral sequences to see whether novel ORF3b variants emerge during the current pandemic.
Associate Professor Kei Sato said that "To our knowledge, this study is the first investigation revealing the role of a SARS-CoV-2-encoded protein that can be associated with the progression of COVID-19".
More information: Yoriyuki Konno et al, SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant, Cell Reports (2020). DOI: 10.1016/j.celrep.2020.108185
Journal information: Cell Reports
Provided by University of Tokyo