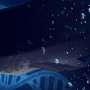
Wels catfish genome assembled

An international research team led by scientists from Estonian University of Life Sciences has for the first time sequenced and assembled the genome of the wels catfish (Silurus glanis). The maximum reported size of the wels catfish is 5 m and up to 300 kg, which makes it one of the largest freshwater fish species in the whole world. By deciphering the genetic code of the barbelled giant, scientists expect to better understand the secrets of the wels catfish's exceptionally rapid growth, enormous appetite and longevity.
The wels catfish lives in large European rivers and lakes. A catfish hunts mainly at night and is not a picky eater, with invertebrates, fish, frogs, rodents and birds in its regular diet. When the water is warm and food is plentiful, the catfish grows extremely rapidly: ten-year-old fish can reach one and a half meters length. Given that a catfish can live up to 80 years, it is no wonder it has rightly become the prized trophy fish and a central character in many legends among anglers.
Due to its rapid growth and tender, boneless flesh, the catfish is increasingly gaining popularity for recreational fishing and aquaculture. The greatest number of wels catfish are being caught from the inland waters of Russia, Kazakhstan and Turkey, and its aquaculture production is currently approximately 2000 tons per year. At the same time, the lack of genetic information in wels catfish has inhibited application of modern selective breeding methods that utilize genomic information instead of phenotypic traits to estimate the breeding values of fish. "The assembled wels catfish genome allows researchers to find genomic regions and gene variants that impact growth rate, age at sexual maturity, disease resistance and other relevant traits for aquaculture," explained Riho Gross, Chair professor of Aquaculture at Estonian University of Life Sciences, who led the research.
According to Anti Vasemägi, senior researcher of Estonian University of Life Sciences and professor of Swedish University of Agricultural Sciences, who participated in the work, the size of wels catfish genome can be compared to that of other bony fishes (800 million base pairs), and contains a little more than 21,000 genes. He added that the genome assembly will serve as a springboard for future research aimed at addressing the bottlenecks in catfish aquaculture and challenges linked to conservation of wild populations.
More information: Mikhail Yu. Ozerov et al, Draft Genome Assembly of the Freshwater Apex Predator Wels Catfish (Silurus glanis) Using Linked-Read Sequencing, G3: Genes|Genomes|Genetics (2020). DOI: 10.1534/g3.120.401711
Provided by Estonian Research Council