Researchers at the University of California, Davis and the MRC Laboratory of Molecular Biology in Cambridge, U.K. have solved the the structure of the complex formed when mRNA is being scanned to find the starting point for translating RNA into a protein. The discovery, published Sept. 4 in Science, provides new understanding of this fundamental process.
"This structure transforms what we know about translation initiation in human cells and there has been a tremendous excitement from people in the field," said Christopher Fraser, professor of molecular and cellular biology at UC Davis and corresponding author on the paper.
Although nearly all our cells contain our entire genome, cells use different subsets of genes to make the proteins they need to perform their various functions. This requires precise control over the processes by which the DNA is first transcribed to produce mRNA and then mRNA is translated to make protein.
Translation begins when a ribosome attaches to a piece of mRNA and scans along it until it finds a start codon, three letters of RNA that say "start translating here." There are over a dozen different proteins known as initiation factors involved in this process. Many of these initiation factors have been found to be dysregulated in various cancers.
However, just how the factors come together and scan mRNA has been poorly understood, due to the lack of understanding of the structures of the entire complex.
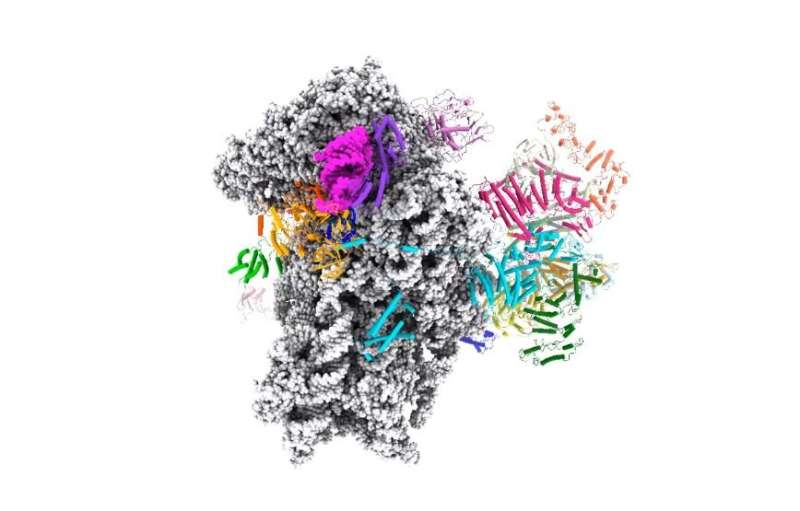
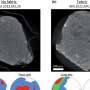
Structure of the human translation initiation complex. Although nearly all our cells contain our entire genome, cells use different subsets of genes to make the proteins they need to perform their various functions. This requires precise control over the processes by which the DNA is first transcribed to produce mRNA and then mRNA is translated during protein synthesis. Researchers at UC Davis and the MRC Laboratory of Molecular Biology have now solved the structure of the protein complex formed when mRNA is being scanned to find the start codon, providing new understanding of the molecular mechanisms underpinning initiation of translation. Credit: Ramakrishnan Lab, Laboratory of Molecular Biology, U.K.
To investigate this, Fraser and postdoctoral researcher Masaaki Sokabe at the UC Davis Department of Molecular and Cellular Biology collaborated with Venki Ramakrishnan, Jailson Brito Querido, Sebastian Kraatz and Yuliya Gordiyenko at the LMB to visualise the structure of the complex. Ramakrishnan shared the 2009 Nobel Prize for Chemistry for his work on the structure of the ribosome.
The team used an mRNA that lacked a start codon so that it would be trapped in the act of scanning. While big for a biological machine, you could fit about 3000 of these complexes across the width of a human hair. The team therefore used cryoelectron microscopy at the LMB to obtain a structure of the complex including the trapped mRNA. Cryoelectron microscopy allows biologists to capture three-dimensional movies of biological molecules down to the scale of single atoms.
Based on this structure, the researchers proposed a model of how the mRNA slots into a channel in the small ribosomal subunit, and a mechanism for how the mRNA might be pulled through the ribosome for scanning, like a strip of film through an old-style projector.
They were able to predict that for most mRNAs, the start codon would need to be sufficiently far from the front end of the mRNA for it to be found in the scanning process, which was the confirmed biochemically by Sokabe and Fraser. Further conformation of the model was obtained by mass spectrometry carried out by Mark Skehel of the LMB.
The UC Davis College of Biological Sciences recently opened its own cryoelectron microscopy facility, which will make this kind of work possible on campus, Fraser said.
More information: "Structure of a human 48S translational initiation complex" Science (2020). science.sciencemag.org/cgi/doi … 1126/science.aba4904
Journal information: Science
Provided by UC Davis