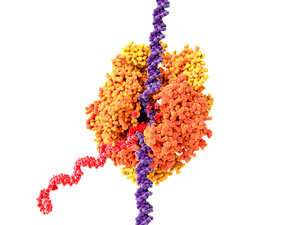
Figure 1: Illustration of DNA (purple) being transcribed to produce messenger RNA (red). RIKEN researchers have discovered the proteins responsible for regulating this transcription in single mouse embryonic stem cells. Credit: Juan Gaertner/Science Photo Library
New insights into what causes neighboring, genetically identical stem cells in mouse embryos to behave differently in terms of the proteins they produce could have implications for regenerative medicine and the early detection of cancer. Cells are like miniature factories that produce proteins that organs need to function. This production process begins with the copying, or transcription, of DNA into messenger RNA (mRNA).
When viewing lots of cells en masse this transcription seems smooth and continuous. But the situation is very different on a single-cell level, where DNA transcription proceeds in fits and starts. This erratic nature of DNA transcription, which is dubbed transcriptional bursting, is partly why cells with identical DNA in the same environment differ from each other. Despite much research, nobody really knows the molecular mechanisms that cause the fluctuation in gene expression in the cells of mammals.
Now, by analyzing the mRNA produced by single embryonic stem cells from mice, Itoshi Nikaido of the RIKEN Center for Biosystems Dynamics Research, Hiroshi Ochiai of Hiroshima University, and co-workers have identified some of the proteins that play a key role in regulating the kinetics of transcriptional bursting. Specifically, they found that a combination of proteins that bind to promoters and gene bodies, including the polycomb repressive complex 2 (PRC2) and transcription elongation factors, control the timing of transcriptional bursting.
This finding goes against a popular theory that hypothesizes that a single 'master regulator' controls the process. "Many researchers believe that the mechanism of transcriptional bursting is controlled by a master regulator," notes Ochiai. "Surprisingly, our data indicates that the kinetic properties of transcriptional bursting of individual genes are not driven by a single master regulator but by a combination of factors that bind to promoters and gene body."
The finding was made possible by a highly reproducible and sensitive method for analyzing mRNA produced by single cells, which was developed by the team in 2018. "This method strongly contributed to quantifying transcription bursting in this study," says Nikaido.
The team anticipates that their fundamental research could find practical applications. "Our findings and techniques may help to establish efficient methods for inducing the differentiation of pluripotent stem cells, such as human iPS cells, into specific cell types," says Nikaido. "Thus, they are expected to be applied to regenerative medicine."
Another possible application is cancer. "Since we know cellular heterogeneity that does not involve genetic mutations occurs when cancer cells grow in the body, we're applying these methods to study the origin and evolution of cancer," says Nikaido. "This research could lead to the development of diagnostic techniques for the early detection of cancer."
More information: Hiroshi Ochiai et al. Genome-wide kinetic properties of transcriptional bursting in mouse embryonic stem cells, Science Advances (2020). DOI: 10.1126/sciadv.aaz6699
Journal information: Science Advances
Provided by RIKEN