Credit: RUDN University
A team of specialists in mathematical modeling from the RUDN University suggested a qualitative model of virus evolution and of the occurrence of new strains. The results of the study can make predicting virus behavior more efficient and help in the development of new antiviral medications. The article was published in the journal Mathematics.
The interaction between a virus and a human body is a very complex phenomenon that can be reduced to two processes: a virus multiplies in host cells, and the organism resists the infection by means of the immune response. A closer look at a virus infection shows that viruses compete for host cells, change under the influence of antiviral drugs, and mutate during its replication. They can evolve at a very high speed by means of modifying old or acquiring new RNA or DNA sequences. Because of these and many other factors, it is difficult for biologists to predict the dynamics of virus evolution, foresee the occurrence of new strains, and assess their potential resistance levels.
The team of Vitaly Volpert, a mathematician from RUDN University, suggested a mathematical model describing the evolution and diversification of virus quasispecies. The model shows a dynamic interaction between the replication, mutation, and elimination of a virus. The results of the work may be further used to predict the occurrence of virus strains capable of avoiding immune recognition and resisting antiviral medications.
"To keep it simple, the model shows us that existing strains evolve in order to reduce competition between viruses, to weaken virus elimination by the immune response, and to become less sensitive to medication. This tendency leads to the occurrence of drug-resistant strains," says Vitaly Volpert.
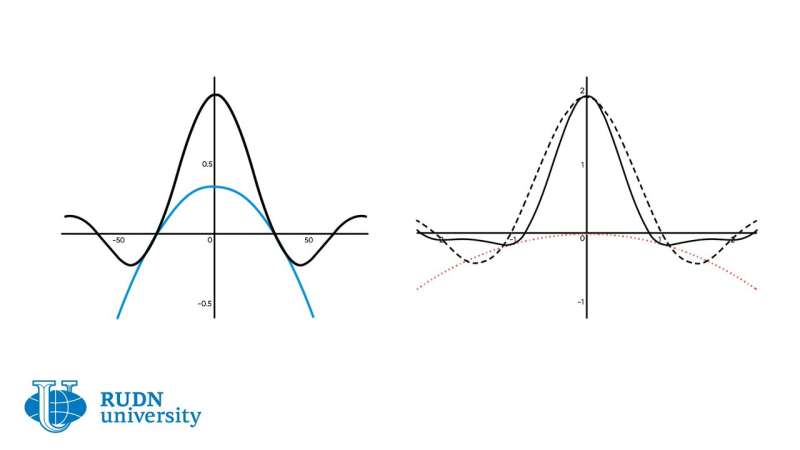
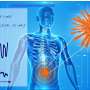
Working together with his colleagues, the scientist described a virus strain as a localized solution concentrated around a given genotype. The emergence of new strains corresponds to a periodic wave propagating in the space of genotypes. The occurrence of new distribution peaks in the process of wave propagation coincides with the occurrence of new virus strains. The team described the conditions of occurrence of periodic traveling waves and their dynamics analyzing the stability of spatially homogeneous stationary solutions.
The model is represented by a nonlocal reaction-diffusion equation for the virus density. The equation has two integral terms that correspond to the nonlocal effects of virus interaction with host cells and immune cells.
The new model is qualitative and applicable to different viral infections. However, to better describe the dynamics of virus quasispecies, one needs to know their individual features, such as the nature of their mutations, interaction with the immune system, and response to antiviral medication.
"Using our model, one can develop various methods to prevent the spread of viral infections across body tissues and the occurrence of new strains, i.e. the propagation in the space of genotypes. However, for these approaches to have a practical implementation, they should be combined with experimental and clinical data," adds Vitaly Volpert.
The model has a number of limitations because it is based on several simplifications. Namely, it does not take into account the existence of the variety of immune cells and cytokines (small informative peptide molecules) that take part in the immune response, or complex processes of intracellular regulation and virus replication. However, these limitations help the scientists to determine some common evolutionary features of virus quasispecies that would be difficult to identify in a more complex model. The work of the team can be used as a basis for further investigations.
More information: Nikolai Bessonov et al. Nonlocal Reaction–Diffusion Model of Viral Evolution: Emergence of Virus Strains, Mathematics (2020). DOI: 10.3390/math8010117
Provided by RUDN University