Host-virus dynamics in a microbial mat in a hot spring microbial mat

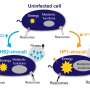
In microbial mats, communities of microorganisms live among viruses that infect them. But what trends govern those virus-host interactions? Do generalist viruses run rampant, capable of infecting different host species? Or, do they tend to specialize, infecting just a single host? Another wrinkle in the mystery is that viruses can affect their prey in different ways. They have the capacity to kill outright, replicating so thoroughly that they lyse the cell—a process called lytic infection. But some viruses are also able to play a long game. In a lifestyle called lysogeny, they can insinuate themselves inside the host's genome, living quietly and replicating with their host indefinitely.
Now, in studying a microbial mat from Cone Pool in Mono County, California, scientists have used sequencing methods for the first time to comprehensively characterize viral-host interactions. The team discovered that the mat's viruses specialize to single hosts, and instead of lysing them, which appears to be prevalent in marine environments, they were predicted to simply abide inside their hosts.
Microbial mats are wildly prevalent, containing an estimated 40 to 80 percent of all the microbial cells on Earth. As microbial predators, viruses are able to determine the turnover of microbial prey, and thus able to influence the metabolic rate of microbial communities. Because microbial metabolism affects the biogeochemical cycling of carbon, nitrogen, and other elements, better understanding the predator-prey dynamics in these mats helps constrain models that estimate biogeochemical fluxes.
Previous work on microbial mats had primarily relied on culturing virus and host pairs in the laboratory to study their interactions. This work, recently published in the ISME Journal, by contrast, relied on methods that eschew culturing, painting a more systematic picture through single-cell and metagenomic sequencing.
A team co-led by Mária Džunková, postdoctoral researcher at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory, used single-cell sequencing to sequence both a cell's genome and detect accompanying viral sequences, which would suggest the virus had been infecting the cell. The team assembled 130 genomes from their sample—a small proportion of the sample's billions of bacterial and archaeal cells—and detected virus in 34 hosts or 26 percent of the single cells.

Metagenomics sequencing then allowed the team to ask, for the 34 hosts they found, how prevalent in the mat are their viruses?
The abundance of a virus relative to its host (inferred through sequence data) can suggest different predatory behaviors. A high relative abundance of virus can suggest a so-called "Kill the Winner" strategy: a virus infects a highly abundant host (a "winner," in terms of numbers), replicates, and lyses its host. Thus, the viral abundance rises as its host's abundance falls.
A low relative viral abundance, on the other hand, suggests that the virus is not rapidly replicating, but instead may be abiding in the host genome: the strategy of lysogeny. The ability of the virus to tag along with the host has earned this strategy a riff, developed by others, on "Kill the Winner"—"Piggyback the Winner."
In the microbial mat, the data suggest viruses are indeed piggybacking their hosts. Though studies in marine environments have detected viruses outnumbering hosts by orders of magnitude, the team found that in the microbial mat, the virus-to-host ratio for most pairs was close to 1. Even the most abundant virus was only 11 times more abundant than its host. There just don't seem to be huge lytic infections happening in this mat.
The team thinks that the piggybacking strategy makes sense in a terrestrial environment with low mobility. The metagenomic data suggests that viruses don't diffuse well through the mat, so they're likely constrained to a local population of host cells. If they were to lyse all those local hosts, they'd likely have a hard time finding more.
More information: Jessica K. Jarett et al. Insights into the dynamics between viruses and their hosts in a hot spring microbial mat, The ISME Journal (2020). DOI: 10.1038/s41396-020-0705-4
Journal information: ISME Journal
Provided by DOE/Joint Genome Institute