Bacterial enzyme extracts rare earth elements in environmentally friendly way

Rare earth elements are vital for many modern technologies. Chemists at LMU have now shown that a cofactor found in a bacterial enzyme can selectively extract some of these metals from mixtures in an environmentally benign fashion.
Rare earth elements (REEs) are an indispensable ingredient of the electronic devices that are now an integral part of our daily lives. They are employed in computers, smartphones, electric motors and many other key technologies as components of magnets and batteries, and also serve as powerful chemical catalysts. REEs comprise 17 elements—scandium, yttrium, lanthanum and the 14 lanthanides that follow lanthanum in the Periodic Table. In nature, they occur as mixtures and are often found in association with the radioactive elements uranium and thorium. All REEs exhibit very similar chemical properties, which makes separating them from each other a difficult, energy-intensive and environmentally problematic task. Now a team led by LMU chemist Professor Lena Daumann has shown that an enzyme cofactor called pyrroloquinoline quinone (PQQ) found in certain species of bacteria selectively binds to specific REEs and can be used to separate them from mixtures.
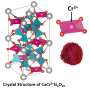
That REEs also play essential roles in the biosphere was discovered less than 10 years ago, when it was shown that certain types of bacteria can selectively take up lanthanides from the environment, which are then incorporated into enzymes for use as metabolic catalysts. For instance, in methylotrophic bacteria, lanthanum or europium bind to PQQ in the enzyme methanol dehydrogenase (MDH), and plays an essential role in the oxidation of methanol—an important part of the energy metabolism of these bacteria. Daumann and her colleagues have now characterized the interaction of PQQ with these REEs in detail and, in cooperation with researchers based in Berlin and Münster, they have isolated PQQ-lanthanide complexes and determined their molecular structures for the first time in the absence of the enzyme matrix.
The results demonstrate that PQQ can selectively remove some REEs by precipitation from aqueous solutions containing mixtures of their salts, without the need for potentially hazardous organic solvents or other additives. Strikingly, PQQ preferentially binds to the larger lanthanides, including neodymium. Recycling of the latter is of particular interest for sustainable technologies. "One characteristic of the lanthanides is that the ionic radius progressively decreases across the row from lanthanum to lutetium, and these minuscule differences can be used to separate them," Daumann explains. Up to now, it was not clear why bacteria preferentially select the larger lanthanides for biochemical functions. Based on their latest results, the authors of the new study suspect that this has to do with the structure of PQQ. Likely the active site in PQQ-containing enzymes has been optimized to accommodate the larger ions in the REE series. The new findings should stimulate further interest in the use of bacteria for the recycling of REEs. The study appears in the journal Chemistry: A European Journal, and is featured on the cover of the latest issue.
More information: Henning Lumpe et al. Cover Feature: The Earlier the Better: Structural Analysis and Separation of Lanthanides with Pyrroloquinoline Quinone (Chem. Eur. J. 44/2020), Chemistry – A European Journal (2020). DOI: 10.1002/chem.202003043
Journal information: Chemistry – A European Journal
Provided by Ludwig Maximilian University of Munich