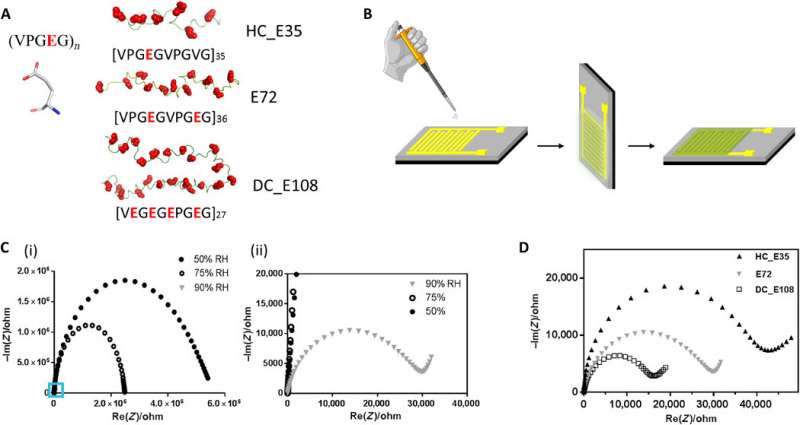
Structure of anionic SUP, devices for proton conductivity measurements and performance of different SUPs in these devices. (A) Primary structure of SUPs engineered with various charge densities. (B) Fabrication protocol of protein films deposited on gold IDEs. (C) Impedance measurement of sample E72 in the shape of Nyquist plot under different RH. The figure (ii) is the zoom-in region of (i) indicated by the blue square. (D) Nyquist plot of protein thin films from samples E72, HC_E35, and DC_E108 equilibrated at RH = 90%. The extrapolated intercept of the observed semicircle with the x axis is indicative of the sample resistance that scales as HC_E35 > E72 > DC_E108. Credit: Science Advances, doi: 10.1126/sciadv.abc0810
Protons are subatomic particles with a positive electric charge. Proton translocation plays a significant role in natural phenomena and manmade technologies. But it remains challenging to control proton conduction and fabrication in biomaterials and devices. In a new report, Chao Ma and an interdisciplinary team of scientists in China, the Netherlands, and Germany, rationally designed proton-conducting protein constituent materials that exceeded previously reported proteinaceous (consisting of or containing protein) systems. They developed the structures through stepwise exploration of peptide sequences from intrinsically disordered coils to protein-supercharged polypeptide chimeras. The new design paradigm offers potential for bioprotonic device fabrication at the interfaces of artificial and biological systems, the results are published on Science Advances.
Proton conduction is responsible for fundamental processes in biology, including bioluminescence, the synthesis of adenosine 5'-triphosphate (ATP) and light-triggered proton translocation. Bioengineers and materials scientists had previously developed several synthetic materials with proton translocation behavior including hybrid systems, although their shortcomings have impeded the fields of bioelectronics and biotechnology. To develop biomaterials dedicated for proton conduction, scientists must explore scaffolds and sequences for their intrinsic proton-conducting behavior. During hydrated states, protons can be transported via water molecules along an adjacent bond network in a mechanism known as proton hopping, which is used as a blueprint to design proton-conducting structures de novo (i.e. from scratch). In this work, Ma et al. developed a stepwise, protein-based proton-conducting membrane with a set of unfolded, anionic supercharged polypeptides (SUPs) containing glutamic acid residues.
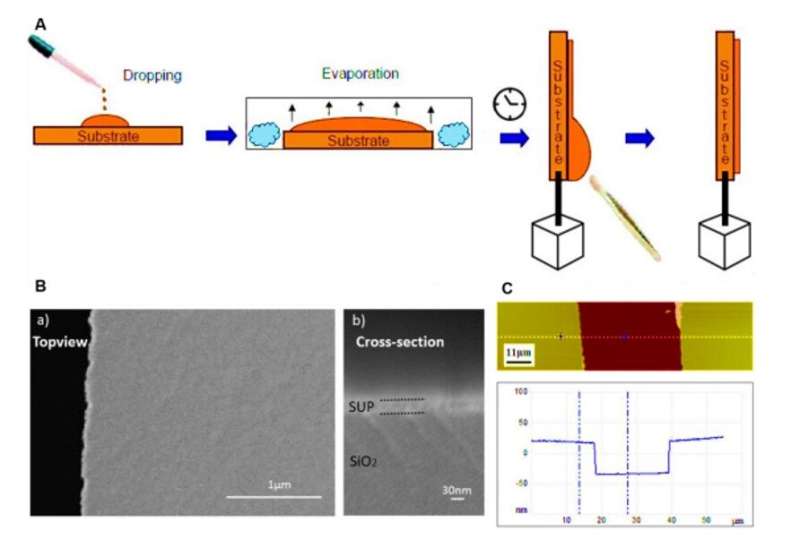
Protein films on substrates and characterization via SEM and AFM. (A) Schematic procedure for the preparation of proton conducting polypeptide and protein films by the drop casting technique used in this study. (B) Scanning electron microscopy (SEM) images showing the flat and homogenous morphology of our customized thin film (here E72 is shown as an example) on the electrodes. The jagged edge on the left side of a) is the truncating position for cross-section imaging in b). (C) AFM image of a scratched thin film surface (top) and its corresponding height profile (bottom). Sample E72 is shown here as an example. Credit: Science Advances, doi: 10.1126/sciadv.abc0810
Developing proton-conducting protein materials
In the polypeptide backbone of the proton-conducting membrane, the hydrophilic (water-loving) charged moieties served as proton carriers. The team studied the proton-conducting performance of these unfolded systems to obtain freestanding membranes and perfected the structural design by amalgamating silk-like β-sheet structures with anionic SUPs to form self-assembled nanostructures. The team decorated the surfaces with dense carboxylic acid groups for hydration, proton dissociation and to form proton conduction pathways. The mechanically stable and freestanding membrane surpassed hitherto reported transport properties of protein-based systems for outstanding proton conductivity.
The team derived the supercharged proteins from elastin; explored previously for applications of protein engineering and interface modification. They introduced glutamic acid (abbreviated Glu or E), which can be easily deprotonated under physiological conditions into the X site of the protein sequence, to form unstructured negatively supercharged polypeptides (SUP-Es). Then they constructed three different variants of supercharged polypeptides known as E72, HC_E35 and DC_E108. Ma et al. used electrochemical impedance spectroscopy (EIS) with gold interdigitated electrodes (IDEs) to evaluate thin-film proton conduction and measured proton transport as a function of relative humidity. When the humidity increased to 90 percent, proton translocation improved due to absorption of a large number of water molecules via the carboxylic acid (-COOH) groups of the material. Besides relative humidity, they also investigated proton conduction relative to charge carrier density for the specimens of interest. By tuning the charge density of the disordered proteins, Ma et al. successfully controlled proton conductance behavior of proteins within films. Due to the high stability and uniformity of the thin films made of SUPs, the setup did not show signs of defects.
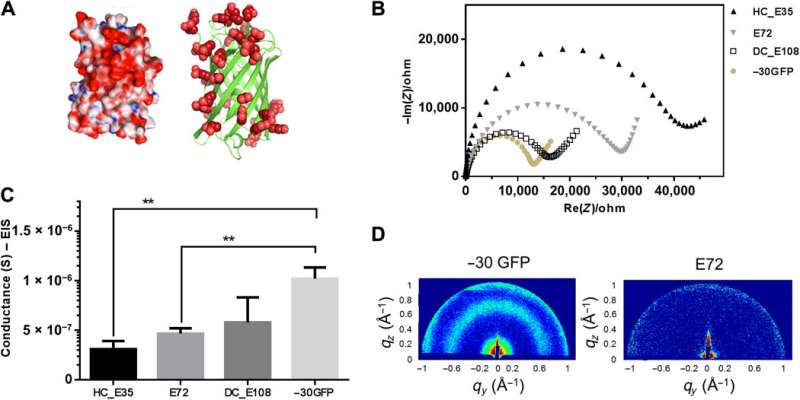
Supercharged −30GFP composed of a nanoscopic β barrel fold for proton conduction. (A) 3D structure of supercharged −30GFP with excessive glutamic/aspartic acids (in red) on the protein surface. The left cartoon visualizes the structure in surface mode, showing positive residues in blue and negative ones in red. The right cartoon visualizes the −30GFP as a ribbon diagram exclusively presenting negative charges. (B) Impedance measurement of sample −30GFP (yellowish solid dots) in Nyquist plotting at 90% RH, compared with other SUP samples. (C) Comparison of conductance between samples E72, HC_E35, DC_E108, and −30GFP (**P = 0.004, n > 3). (D) GIXD patterns for structure investigation of the different films. Two distinct signals were observed for the nanostructured −30GFP (left), while no signal was detected for E72 films (right), indicating its unstructured nature. Credit: Science Advances, doi: 10.1126/sciadv.abc0810
Folded protein backbones and spider-E development
Ma et al. then further studied folded nanosized protein backbones and equipped the nanoscale scaffolds with carboxylic acid on the surface—similar to SUPs. Using X-ray diffraction, they investigated structural information inside the supercharged protein samples to obtain distinct signatures of their structural domains, to show how nanostructured components could facilitate proton translocation. The work allowed the team to rationally engineer protein motifs to perform proton conduction. Motivated by increased proton conductivity, Ma et al. combined the resulting design elements with the existing supercharged polypeptide (SUP) structures.
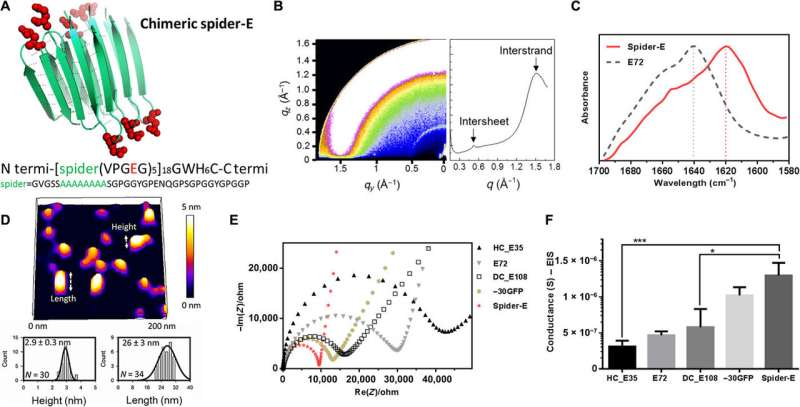
Instead of using β-barrel motifs in the materials architecture, they used the mechanically stable β-sheet structures—a sequence obtained from spider silk. They named the combined system of anionic SUP with β-sheet sequences as 'spider-E'. The scientists produced the recombinant anionic spider-E material using plasmid-vector expression systems in the lab and determined the structure using X-ray diffraction, Fourier transform infrared (FTIR) spectroscopy and atomic force microscopy. The spider-E film showed higher proton conductance compared to amorphous SUP films alone.
Sequence, structure, and proton conduction of recombinant supercharged spider-E thin films on IDE. (A) Rationally designed supercharged spider silk–inspired proteins (spider-E). The spider motif contains a poly-A sequence (green) and anionic supercharged regions (red) that are forming the loops between the rigid β sheets. (B) Structure analyses of the spider-E supported film. Two peaks were detected by GIXD, indicating the characteristic intersheet and interstrand distances, respectively. (C) FTIR characterization of the films indicate random coils for the E72 sample (gray dashed line) with an amide I peak located at 1640 cm−1 and a shift to a typical β sheet amide I peak for the spider-E sample (red solid line) at 1620 cm−1. (D) Morphology analyses of the spider-E supported film. Quantification of the nanostructures assembled through spider β sheet domains by AFM. This sample was obtained by extensive swelling of the film by water contact to induce separation between the domains. (E) Nyquist plots obtained at RH = 90% for the five genetically engineered samples including spider-E. The impedance curve of the spider-E sample shows the lower resistance value among all the samples (red). (F) Comparison of conductance of the resulting devices demonstrating the stepwise increase in the transport properties due to the improved protein design. The proton transport of spider-E thin films on IDEs is noticeably higher than HC_E35 (***P = 0.0009, n > 3) and DC_E108 (*P = 0.0155, n > 3). Credit: Science Advances, doi: 10.1126/sciadv.abc0810
Characterizing the spider-E material architecture
The β-sheet structured material system showed improved mechanical properties as a free-standing membrane that could be easily produced. For instance, Ma et al. drop casted the spider-E solution to engineer a transparent macroscopic membrane in the lab. The results showed mechanical robustness of the construct due to the inclusion of spider motifs with a yield strength comparable to recombinant spider silk materials. The researchers showed how spider motifs formed β-sheet structured domains with hydrophilic surfaces composed of glutamic acid-rich SUP strands, to facilitate excellent proton hopping. The study pushed the limits of existing proteinaceous proton-conducting materials to represent a key example of protein engineering. The work represents one of the first examples that combines protein engineering and the rational design of bulk architecture with collective properties from molecular ensembles.
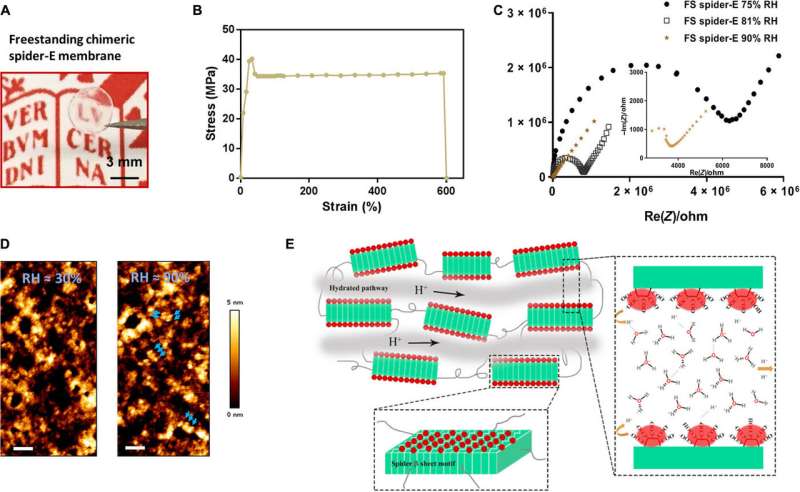
Bulk freestanding chimeric spider-E membrane with extraordinary proton transfer properties. (A) A digital photograph illustrates the dimensions and transparency of the membrane. The protein membrane is clamped with a fine tweezer. Photo credit: Chao Ma, University of Groningen. (B) Mechanical characterization of the freestanding (FS) protein membrane, showing a typical tensile stretching curve. (C) Nyquist plot illustrating the conductance behavior of the FS spider-E membrane under different RHs. The film shows best proton translocation properties at 90% RH. (D) AFM characterization of the FS spider-E membrane under ~30 and ~90% RH conditions. Scale bars, 100 nm. Blue arrows point at distinguishable nanostructures. (E) Proposed mechanism of proton transport in the spider-E membrane at RH = 90%. The protons hop between water molecules nanoconfined in the hydrated network of nanodomains formed by spider β sheet motifs (in green). The glutamic acid residues in the chimeric nanostructures present carboxylic groups (in red) on the surface, providing the protons and coordinating water molecules. Credit: Science Advances, doi: 10.1126/sciadv.abc0810
In this way, Chao Ma and colleagues applied rational molecular de novo design and engineering to achieve a bioinspired protein-derived bulk material with robust properties of proton conduction and excellent mechanical stability. They tested the surface modifications using a range of biophysical tools. The team developed the new generation, bioinspired bulk material and explored successive sequence designs to offer a promising platform for applications in biotechnology and envision the use of such materials for proton transport in miniaturized biofuel cells of the future.
More information: Chao Ma et al. De novo rational design of a freestanding, supercharged polypeptide, proton-conducting membrane, Science Advances (2020). DOI: 10.1126/sciadv.abc0810
I. N. Watt et al. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria, Proceedings of the National Academy of Sciences (2010). DOI: 10.1073/pnas.1011099107
Jeff A. Hurd et al. Anhydrous proton conduction at 150 °C in a crystalline metal–organic framework, Nature Chemistry (2009). DOI: 10.1038/nchem.402
Journal information: Science Advances , Proceedings of the National Academy of Sciences , Nature Chemistry
© 2020 Science X Network