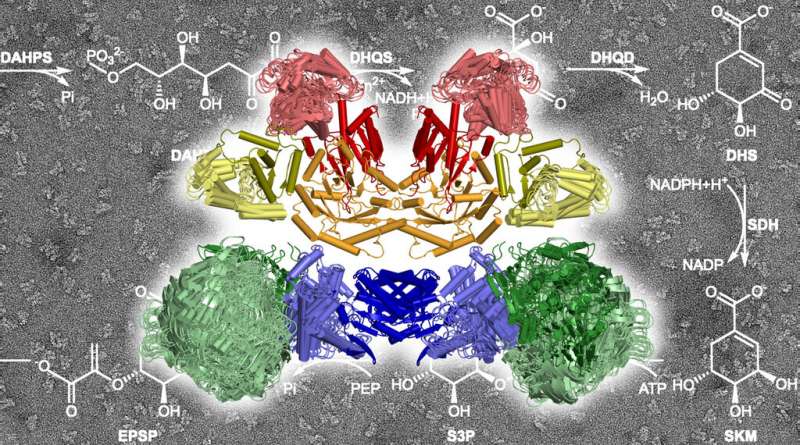
The ten enzymatic components of the AROM complex at work, catalyzing the chemical reactions sketched in the background. Credit: Max Planck Society
For protein molecules that contribute to metabolism, interactions with other components of their metabolic pathway can be crucial. Scientists at the Max Planck Institute for Developmental Biology in Tübingen have now investigated a natural enzyme complex that comprises 10 enzymes with five distinct activities. They found that the molecular architecture is surprisingly compact, yet offers individual enzymes maximum free moving space, which opens up novel perspectives for drug discovery. The scientists have published their results in Nature Chemical Biology.
In textbooks, metabolic pathways always look a bit like assembly line work. One enzyme follows another like pearls on a string. Intermediate products are removed or passed on from one station to another. "However, in the cell, this often does not happen in such an orderly fashion," says Marcus Hartmann, group leader at the Max Planck Institute for Developmental Biology. "The individual components of a metabolic pathway can be located in different areas within the cell, and in most cases, it is not understood, if and how they come together to form ordered structures."
To understand the function and dynamics of metabolic pathways, scientists also look into the interaction and hierarchies of the individual components. This is the only way to grasp the overall picture.
With this in mind, Hartmann and his team have examined an otherwise well-investigated shikimate pathway. It occurs in plants and microorganisms, including pathogenic fungi and parasitic protists such as the pathogens that cause toxoplasmosis or malaria. One of its tasks is to synthesize the precursors of the amino acids phenylalanine, tyrosine and tryptophan, which are essential in animals and humans. This pathway has recently also gained attention as the target of the notorious herbicide glyphosate.
The researchers from Tübingen noted that while the majority of enzymes of the shikimate pathway are present individually in plant and bacterial cells, in fungi and protists, five of the seven components are combined in a large enzyme complex known as AROM. "We wanted to know how this large complex is structured and what the advantages of its architecture might be," says Hartmann. "For example, if catalysis is more efficient in this complex."
The scientists succeeded in solving a crystal structure of the entire protein complex. They found that the five enzyme components, each in two copies, assemble into a compact structure with a total of 10 components in a very small space. "We did not expect such a compact architecture," says Hartmann. "Many enzymes of the shikimate pathway actually need quite a lot of elbow-room for their work."
Thanks to its ingenious architecture, however, the available space in the AROM complex is sufficient. Hartmann says that although the crystal structure only represents a static snapshot of the complex, "the individual components are so well studied that we can simulate in a computer model how they move within the complex."
It turned out that the necessary liberty of action for the individual enzymes is tailored such that they can move independently without collisions. "We actually do not have evidence for any coordination of the individual enzyme motions," says Harshul Arora Verasztó, first author of the study. "It is more like organized chaos."
Further investigations have shown that a mere clustering of enzymes—to form the AROM complex—is not sufficient not provide a catalytic advantage for the shikimate pathway of fungi and protists—at least on the basis of the present data. "A comparison under laboratory conditions has shown that the throughput of the complex is not higher than that of the scattered individual enzymes," says Hartmann. "For biotechnologists who are trying to optimize the efficiency of catalytic cascades via the clustering of enzymes, this might be surprising news."
In the first place, the results obtained by the Tübingen scientists contribute to basic research—to gain insights into how enzymes and in particular enzyme complexes function in metabolic pathways. However, they also open up completely new perspectives for drug discovery. "In this regard, the tight arrangement of the particularly flexible enzyme components within the AROM complex is highly interesting," explains Hartmann.
In most cases, drugs attack at the catalytic centers, at the sites where enzymes perform their reactions. "For the AROM complex, it is conceivable to develop drugs that act like a wedge between the enzymatic components to block them." This approach could lead to the development of inhibitors that only work on fungi or protists, such as the pathogens causing toxoplasmosis or malaria, but do not impair the shikimate pathway of plants and bacteria.
More information: Harshul Arora Verasztó et al. Architecture and functional dynamics of the pentafunctional AROM complex, Nature Chemical Biology (2020). DOI: 10.1038/s41589-020-0587-9
Journal information: Nature Chemical Biology
Provided by Max Planck Society