Scientists uncover pivotal virus structure for replication
Researchers from the Monash Biomedicine Discovery Institute (BDI) have revealed the structure of a protein complex that is essential for the poliovirus to replicate. The structure may be similar to those in other RNA viruses and form a potential target for the development of antiviral drugs halting virus replication more broadly.
Scientists in Professor Jackie Wilce and Professor Matthew Wilce's laboratory used poliovirus—representative of the picornavirus family of viruses that includes the virus causing the common cold—as their virus of focus.
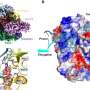
The poliovirus delivers RNA into our cells that contains the code for making more copies of itself. To do this it needs to trick our cellular machinery (the ribosome) into docking at one end, allowing it to read the RNA sequence to make viral proteins. The researchers used cryo-electron microscopy at Monash and other biophysical methods to show the way in which the PCBP2 protein binds to stem loop IV of the poliovirus' internal ribosome entry site (IRES) allowing the ribosome to dock in the first stage of viral RNA translation.
"It was already known that the protein binding to this part of the poliovirus RNA was absolutely essential for the ribosome to be able to dock the RNA but nobody really understood what the 3-D shape was," Professor Jackie Wilce said.
The structure had been of interest to scientists for many years with researchers worldwide slowly building an understanding of the mechanism involved; this study marked the first time it was seen in 3-D.
"This is like the icing on the cake to be able to see it in three dimensions," she said. "It's always amazing to be the first to visualize a molecular structure that no eyes have ever set on before."
The study's findings were important for understanding the biology involved but may also spark new ideas to develop a novel class of antiviral therapeutics that interrupt viral replication, Professor Jackie Wilce said. The researchers will now study the biological process further and are also testing ideas about inhibitors to interrupt it.
The study, conducted with collaborators at the BDI and poliovirus expert Professor Bert Semler from the University of California Irvine, was published in Nucleic Acids Research.
Drs Simone Beckham and Mehdi Matak are the joint first authors in the study with Professors Jackie and Matthew Wilce joint senior authors. They received expert input from Dr. Matthew Belousoff, Hari Venugopal and Dr. Hans Elmund for deriving the cryo-EM structure. Vital contributions were also made by other Wilce lab members along with Professor Bert Semler and Dr. Joseph Nguyen. The research was supported by the Australian Synchrotron for the collection of some data.
More information: Simone A Beckham et al. Structure of the PCBP2/stem–loop IV complex underlying translation initiation mediated by the poliovirus type I IRES, Nucleic Acids Research (2020). DOI: 10.1093/nar/gkaa519
Journal information: Nucleic Acids Research
Provided by Monash University