Comparing 13 different CRISPR-Cas9 DNA scissors
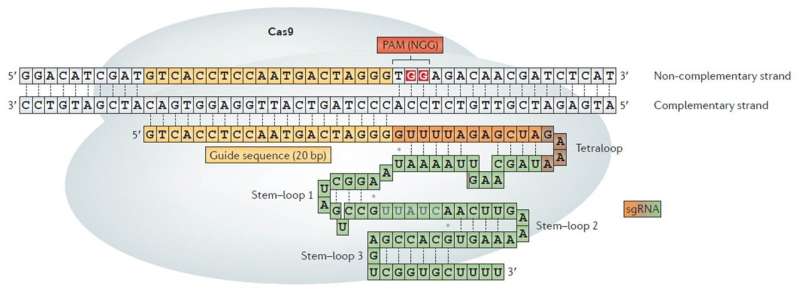
CRISPR-Cas9 has become one of the most convenient and effective biotechnology tools used to cut specific DNA sequences. Starting from Streptococcus pyogenes Cas9 (SpCas9), a multitude of variants have been engineered and employed for experiments worldwide. Although all these systems are targeting and cleaving a specific DNA sequence, they also exhibit relatively high off-target activities with potentially harmful effects.
Led by Professor Hyongbum Henry Kim, the research team of the Center for Nanomedicine, within the Institute for Basic Science (IBS, South Korea), has achieved the most extensive high-throughput analysis of CRISPR-Cas9 activities. The team developed deep-learning-based computational models that predict the activities of SpCas9 variants for different DNA sequences. Published in Nature Biotechnology, this study represents a useful guide for selecting the most appropriate SpCas9 variant.
This study surpassed all previous reports, which had evaluated only up to three Cas9 systems. IBS researchers compared 13 SpCas9 variants and defined which four-nucleotide sequences can be used as protospacer adjacent motif (PAM) – a short DNA sequence that is required for Cas9 to cut and is positioned immediately after the DNA sequence targeted for cleavage.
Additionally, they evaluated the specificity of six different high-fidelity SpCas9 variants, and found that evoCas9 has the highest specificity, while the original wild-type SpCas9 has the lowest. Although evoCas9 is very specific, it also shows low activity at many target sequences: these results imply that, depending on the DNA target sequence, other high-fidelity Cas9 variants could be preferred.
-
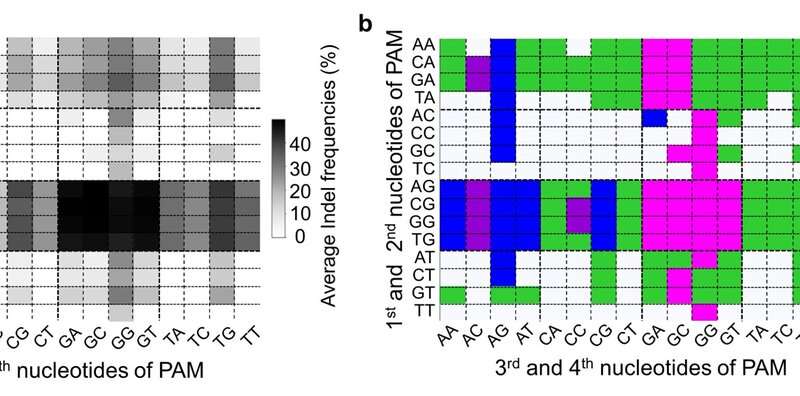
Figure 2. PAM compatibilities for SpCas9 variants. (a) Darker colors indicates higher frequency of DNA cleavage. (b) Among these four variants (SpCas9, VRQR, xCas9 and SpCas9-NG), SpCas9-NG has been the traditional choice for all PAM sequences that have a guanine (G) as the second nucleotide. However, these results shows that for PAM sequences AGAG and GGCG, for example, the Cas9 variant VRQR (in blue) would be preferable. Credit: Institute for Basic Science -
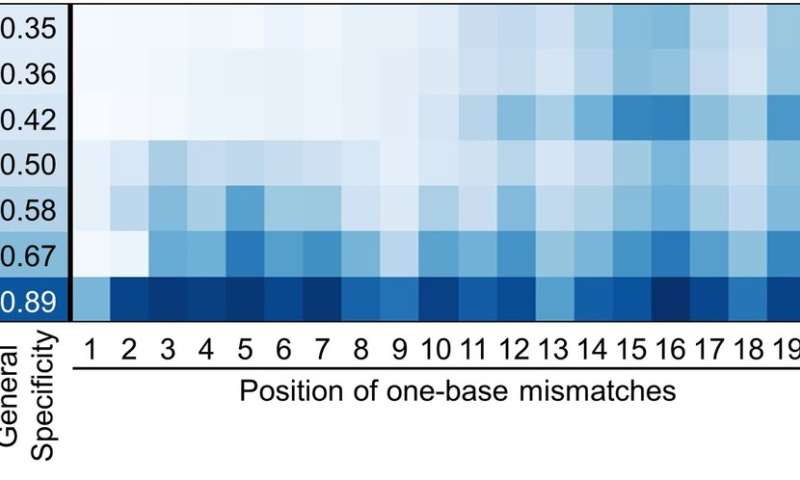
Figure 3. Comparing the specificity of the SpCas9 variants with a DNA sequence that has a single mismatch between the guide RNA and the target sequence. evoCas9 and the original SpCas9 exhibit the highest and the lowest specificity, respectively. Credit: Institute for Basic Science
Based on these results, IBS researchers developed DeepSpCas9variants (deepcrispr.info/DeepSpCas9variants/), a computational tool to predict the activities of SpCas9 variants. By accessing this public website, users may input the desired DNA target sequence, find out the most suitable SpCas9 variant and take full advantage of the CRISPR technology.
"We began this research when we noticed the critical lack of a systematic comparison among the different SpCas9 variants," says Kim. "Now, using DeepSpCas9variants, researchers can select the most appropriate SpCas9 variants for their own research purposes."
More information: Nahye Kim et al. Prediction of the sequence-specific cleavage activity of Cas9 variants, Nature Biotechnology (2020). DOI: 10.1038/s41587-020-0537-9
Journal information: Nature Biotechnology
Provided by Institute for Basic Science