Epithelial GPS: Position of RNAi machinery is associated with epithelial identity
Epithelial cells are held together and connected by several different types of structures that form cell-cell contacts. One of these structures, found near the top surface of the cell, is the adherens junction. This junction is critical for organ development, tissue architecture and cell function; disruption of adherens junctions can lead to inflammatory bowel disease, disorders of the skin and hair, and certain types of cancer.
Researchers at the Medical University of South Carolina (MUSC) who study adherens junctions have identified the RNA interference (RNAi) machinery, a biological process that regulates gene expression, as a critical interacting complex that influences epithelial cell biology. Their results, published on Apr. 7 in the International Journal of Molecular Sciences, showed that the RNAi machinery is predominantly localized at adherens junctions in normal colon epithelial cells but is mis-localized in colon cancer.
"This goes against the dogma in the field that the core components of the RNAi machinery localize either in the nucleus or the cytoplasm. When you look at tissues, it is so dominant at the apical junctions and a very strong feature of the colonic epithelium," said Antonis Kourtidis, Ph.D., an assistant professor in the department of Regenerative Medicine and Cell Biology and associate member of Hollings Cancer Center, who studies adherens junctions in cell behavior and disease.
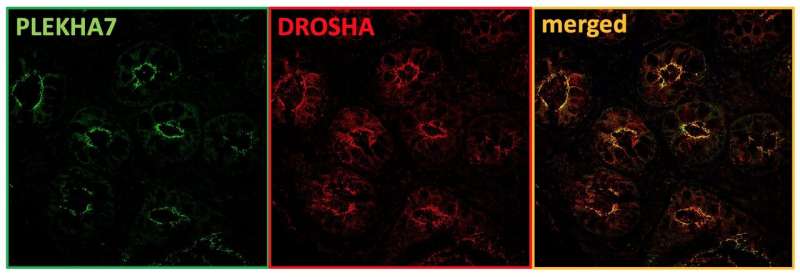
The Kourtidis Lab focuses on the intersection of cell-cell interactions and RNA biology. Previous work from Dr. Kourtidis has shown that in Caco2 cells (a human intestinal epithelial cell line), the adherens junction protein PLEKHA7 (pleckstrin homology domain containing A7) recruits numerous RNA-binding proteins including members of the core RNAi machinery. At the adherens junction, the RNAi machinery regulated the processing and function of tumor suppressing microRNAs (miRNAs). Importantly, when PLEKHA7 was depleted, the RNAi machinery was mis-localized, several of these miRNAs were dysregulated, and the cells became more tumorigenic.
The Kourtidis Lab decided to build on these past findings in this current paper. "How physiologically relevant is this to either normal tissues or to disease," asked Kourtidis. "In this paper, we wanted to see what holds true for the colonic epithelium."
Examination of primary colonic epithelial cells and of normal colon tissues confirmed that the RNAi machinery co-localized with PLEKHA7 at adherens junctions; however, both PLEKHA7 and the RNAi machinery were mis-localized in human tumor samples at all stages of disease. Junctional localization of PLEKHA7 and the RNAi machinery was also lost in several human colon cancer cell lines. Interestingly, it was noted that PLEKHA7 in these cell lines was mis-localized due to increased activity of the human kinase Src, a well characterized proto-oncogene and promoter of colon cancer. Pharmacological inhibition of Src restored junctional localization of PLEKHA7 and the RNAi machinery in one of the human colon cancer cell lines, hinting at a potential mechanism of regulation.
Building on that potential mechanism, the Kourtidis lab examined another human colon cancer cell line in which PLEKHA7 was downregulated. Turning on expression PLEKHA7 in these cells restored localization of the RNAi machinery at the adherens junction, increased the levels of tumor suppressing miRNAs, and suppressed tumor growth in a mouse model. Together, these data suggest that PLEKHA7 and the junctional localization of the RNAi machinery may play a tumor suppressive role in epithelial cells.
"We still haven't nailed the tumor suppressing function. Every indication that we have so far suggests that this is the case, but we really need to confirm this in more specific mouse models," said Kourtidis.
In summary, localization of the RNAi machinery to adherens junctions is a predominant feature of healthy colon epithelium. Furthermore, the localization and activity of the RNAi machinery is disrupted in colon cancer. Kourtidis pictures this mechanism acting like a guardian, at least to some extent, of epithelial homeostasis.
Kourtidis says there's still a lot left to learn, but he's looking forward to it. A major question that remains is determining how this system is regulated. Because Src inhibition restored localization of the RNAi complex only in some cancer cells but not others, the Kourtidis lab is looking at different mechanisms regulating the localization of PLEKHA7 and the RNAi machinery. Furthermore, looking downstream at how the miRNAs that are dysregulated when the system is perturbed contribute to disease progression is an essential question that needs to be answered.
Interestingly, PLEKHA7 has been shown to be downregulated or mis-localized in human breast and kidney tumors, and Kourtidis thinks that this is a phenomenon important for epithelial homeostasis. Epithelial cells can lose their identity when the RNAi machinery is lost or mis-localized, and restoration of the RNAi machinery localization (or the downstream miRNAs) could serve as a brake on tumor progression.
More information: Joyce Nair-Menon et al, Predominant Distribution of the RNAi Machinery at Apical Adherens Junctions in Colonic Epithelia Is Disrupted in Cancer, International Journal of Molecular Sciences (2020). DOI: 10.3390/ijms21072559
Provided by Medical University of South Carolina