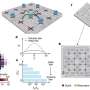
Overview of the cultivated alfalfa genome. Credit: Nature Communications
Improvement of cultivated alfalfa (Medicago sativa), a perennial herbaceous legume and one of the most important forage crops, might be accelerated if agronomically beneficial mutations could be easily incorporated into modern varieties. However, obtaining mutants of the autotetraploid and self-incompatible cultivated alfalfa through traditional phenotypic selection is challenging. No mutant of the species has been previously reported using either CRISPR/Cas9 or other site-specific nucleases.
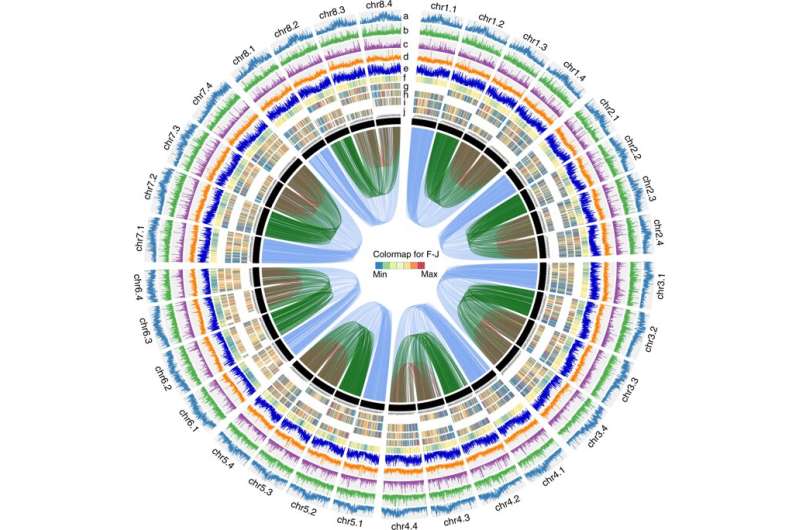
In a study published in Nature Communications, researchers applied PacBio CCS (circular consensus sequencing) and Hi-C (High-throughput chromosome conformation capture) technology to generate an allele-aware chromosome-level genome assembly for the cultivated alfalfa consisting of 32 allelic chromosomes.
On the basis of the genome assembly, the researchers also developed an efficient CRISPR/Cas9-based genome editing protocol and precisely introduced tetra-allelic mutations into null mutants that displayed obvious phenotype changes.
Most importantly, the mutated alleles and phenotypes can be stably transmitted to progenies by cross-pollination between two mutants in a transgene-free manner, which may help to accelerate the breeding speed and mitigate concerns about transgene technology and its products.
"By exploiting new sequencing technology and Hi-C scaffolding, we are able to decode the complex autotetraploid cultivated alfalfa genome, reveal events that have apparently shaped it, and create foundations for further studies on legumes and complex genome assembly," said Prof. Chen Jianghua, one of the three correspondence authors of the study.
"Our results also provide robust foundations for further technical developments, which could potentially raise global food security by reducing breeding periods and costs of improving key agronomic traits of this important crop," added Chen Jianghua.
More information: Haitao Chen et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa, Nature Communications (2020). DOI: 10.1038/s41467-020-16338-x
Journal information: Nature Communications
Provided by Chinese Academy of Sciences