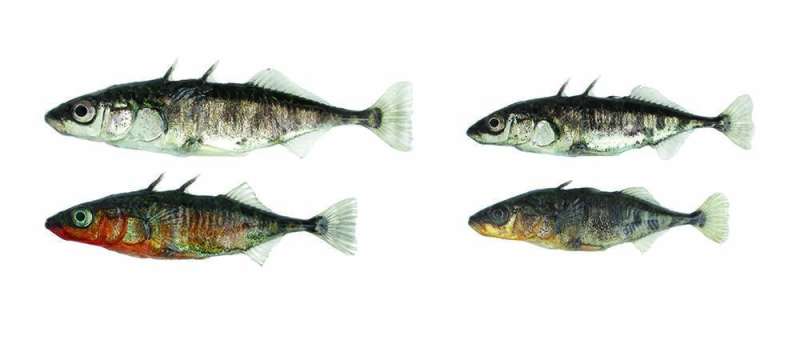
Two ecotypes of threespine stickleback fish can be found in and around Lake Constance. Each has developed under the influence of its specific habitat: lake stickleback on the left and river stickleback on the right. The two ecotypes differ in numerous morphological and behavioral traits; most striking are the differences in body size and in the breeding coloration of the males (lower row). Credit: University of Basel, Daniel Berner
Researchers from Basel have identified the genetic basis of rapid adaptation using a native fish species. They compared threespine stickleback fish from different habitats in the Lake Constance region. Their study reveals that changes in the genome can be observed within a single generation. The results were published in the journal Nature Communications.
Evolution is usually viewed as a slow process, with changes in traits emerging over thousands of generations only. Over the recent years, however, research has indicated that adaptation in specific traits can occur more quickly. However, very few studies outside microorganisms were able to demonstrate empirically how quickly natural selection shapes the whole genome.
A research team led by Dr. Daniel Berner at the University of Basel's Department of Environmental Sciences has now provided evidence for rapid evolution within a single generation, using threespine stickleback fish as model organism. The five-year study combined lab work, field experiments, mathematical modeling and genomic analysis.
Different habitats: lakes and rivers
In the Lake Constance area, stickleback have adapted to ecologically different habitats—lakes and rivers. To examine how quickly adaptation occurs across the genome, lake- and river-dwelling fish were crossed in the laboratory over several generations. The genomes of the two ecotypes were thus mixed, resulting in a genetically diverse experimental population.
The researchers released 3000 experimental fish into a natural river habitat without stickleback, exposing them to natural selection. After one year, the remaining fish were recaptured and examined genetically. Credit: University of Basel, Dario Moser
In a second step, the researchers released thousands of these experimental fish into a natural river habitat without resident stickleback, exposing them to natural selection. After a year, the remaining fish were recaptured and examined genetically.
"The hypothesis of this experiment was that in the river habitat in which the experimental animals had to survive, genetic variants of the original river population would increase in frequency," says Berner. "However, we had no idea whether this would be measurable within a single generation."
Genomic analysis confirms hypothesis
To record potential changes in the genome, the researchers first had to identify the DNA regions most likely to be targeted by natural selection. To do so, they compared the original lake and river populations based on DNA sequence data. This revealed hundreds of regions in the genome likely important for adapting to the lake and river conditions. In precisely these regions, the experimental population's DNA sequence data from before and after the field experiment were then compared to identify changes in the frequency of genetic variants.
The result supported the hypothesis: on average, the frequency of the river variants increased by around 2.5% at the expense of the lake variants. "This difference might appear small at first glance, but is truly substantial when extrapolated over a few dozen generations," says Berner. The experiment demonstrates that evolution can occur very quickly right in front of our eyes—and not only in microorganisms. "Such rapid evolution may help some organisms to cope with the current rapid environmental changes caused by humans," Berner concludes.
More information: Genomic release-recapture experiment in the wild reveals within-generation polygenic selection in stickleback fis. Nature Communications. (2020), DOI: 10.1038/s41467-020-15657-3
Journal information: Nature Communications
Provided by University of Basel