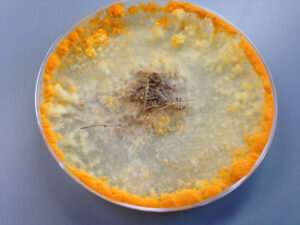
The filamentous fungus Neurospora crassa eating plant biomass. Credit: Vincent Wu
A team led by researchers at the University of California, Berkeley used a multi-omics approach to reconstruct and model gene regulatory pathways used by the filamentous fungus Neurospora crassa, and to identify and decide on the order in which this fungus breaks down plant cell wall materials including lignin, cellulose and hemicellulose.
N. crassa is the model organism for filamentous fungi, all of which contain a wide array of plant cell-wall degrading enzymes (PCWDEs) that allow them to efficiently break down the wide array of simple and complex components present in plant biomass. This is of interest for bioenergy researchers looking to improve the industrial production of sustainable biofuels and bioproducts. Filamentous fungi are also being used in the biotechnology industry to produce enzymes, proteins, and other chemicals.
Filamentous fungi are like handymen who show up at a job site for a task that requires a flathead screwdriver with a full toolbox including Phillips and specialty screwdrivers, not to mention Allen wrenches. The fungi are similarly armed with a variety of PCWDEs to first break down the components of plant cell walls, which range from simple to complex carbohydrates, and then convert them into simple sugars. More importantly, when faced with a veritable buffet of carbon sources, these fungi detect which complex chains are available; this information triggers pathways to determine which enzymes should be deployed in what order to most efficiently degrade the plant biomass.
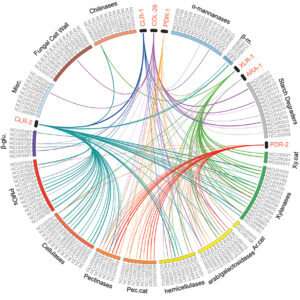
This figure shows overlapping regulation by the transcription factors responsible for turning on enzymes for digesting the plant cell wall. The lines connect transcription factors to degradative enzymes that the factors transcriptionally control. Many enzymes can be controlled by multiple transcription factors, and might explain the capability of filamentous fungi to fine tune expression of given enzymes given a particular resource. Credit: Wu et al, PNAS 2020
In order to learn more about these regulatory networks in the model fungus N. crassa, a team led by N. Louise Glass at the University of California, Berkeley and her postdoctoral fellow Vincent Wu worked with researchers at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab). The work was enabled in part through JGI's Community Science Program through a proposal akin to the Encyclopedia of DNA Elements (ENCODE) project, which is aimed at determining the activity, expression and regulation of protein-coding genes.
This first paper integrating multi-omics data from this JGI fungal ENCODE proposal was recently published in the Proceedings of the National Academy of Sciences. Researchers applied multiple omics techniques to reconstruct and model the gene regulatory networks as they responded to available carbon sources, which ranged from simple sugars to plant biomass. One of these techniques was DAP-seq, a high-throughput method for identifying protein binding sites in DNA by DNA Affinity Purification, developed and optimized by Ronan O'Malley, who leads the JGI's Sequencing Technologies group. DAP-seq allowed the team to identify the direct binding sites of many transcription factors to better understand how nutrients are acquired and carbon is metabolized in filamentous fungi. The work also suggests a new approach to gene annotation, which then can be explored in depth for multiple models and their close relatives across the fungal tree of life, enriching the work being done as part of the 1000 Fungal Genomes Project.
More information: Vincent W. Wu et al. The regulatory and transcriptional landscape associated with carbon utilization in a filamentous fungus, Proceedings of the National Academy of Sciences (2020). DOI: 10.1073/pnas.1915611117
Journal information: Proceedings of the National Academy of Sciences
Provided by DOE/Joint Genome Institute