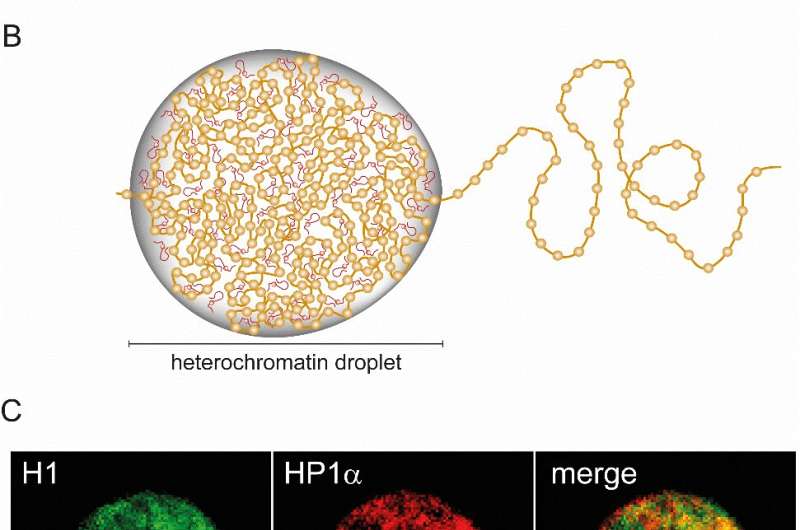
(A) Cartoon depiction of chromatin packaged into compact heterochromatin and loosely packed euchromatin. (B) The segregation of heterochromatin and euchromatin is mediated by liquid-liquid phase separation with linker histone H1. (C) Two-color fluorescent microscopy images of histone H1 and HP1α (a heterochromatin marker protein) showing co-localization of H1 and heterochromatin in liquid-like droplets within HeLa nuclei. Credit: Institute for Basic Science
How can human cells pack 3-meter-long DNA into their tiny nuclei and unpack it only where and when it is needed? This fascinating process is far from being completely understood.
Researchers at the Center for Soft and Living Matter, within the Institute for Basic Science (IBS, South Korea), made a significant advancement in our understanding of the organization of genomic DNA within the nucleus. These findings, published in the Biophysical Journal, describe a new function of histones, which are proteins responsible for packaging genomic DNA and are involved in gene regulation.
DNA wraps around a complex of four core histones (H2A, H2B, H3, and H4), forming a structure known as the nucleosome core particle (NCP). Another histone, the linker histone H1, binds outside of the NCP and links the NCPs together to form a tightly packed structure. This packaging, however, varies during the lifetime of the cell. Cells capable of dividing spend most of their time in interphase, during which they are actively producing proteins and duplicating their DNA before undergoing cell division. During this stage, DNA in complex with histone proteins appears in a "loosened up" state. Its organization, however, is not as random as it might look: specific genes group together depending on their function and occupy specific locations within the nucleus. Genes that are active are grouped together in loosely packed domains called euchromatin, while genes that are silenced are partitioned into densely packed domains called heterochromatin.
Recently, a concept has emerged that formation of heterochromatin is driven by liquid-liquid phase separation (LLPS). LLPS is a physical phenomenon observed in vitro when oppositely charged polymers, mixed in solution, partition into droplets that have a high concentration of both polymers. In the last decade, this phenomenon has excited biologists as it is becoming more and more evident that LLPS plays an important role in forming membrane-less compartments as a way to organize the cell interior. The team realized that histones share many characteristics with other proteins that had been shown to undergo LLPS, namely they contain extended intrinsically disordered regions and have an excess of charged residues.
The researchers discovered that the linker histone H1 is involved in LLPS of heterochromatin in the human cell nucleus. "About three years ago, we had started experimenting with histones and DNA oligomers in vitro and were very excited to find that the DNA formed droplets with a mixture of histones, and that the histones were highly mobile within the droplets. In particular, H1 alone was the best at forming liquid droplets," says Anisha Shakya, lead and corresponding author of the study. "This seeded our interests to study H1 in cells; to test the hypothesis that H1 undergoes LLPS in cells, in particular in the context of heterochromatin formation, where it is found in elevated concentrations."
By culturing HeLa cells expressing fluorescently tagged H1, the research team imaged the spatial and temporal distribution of this protein during the interphase of the cell cycle. Fluorescence microscopy showed H1 localized in small puncta in the nucleus that would occasionally merge with each other: a sign that they are liquid-like. Other experiments confirmed that DNA was densely packed within these droplets, and was enriched in the protein HP1α, indicating heterochromatin regions. Interestingly, in vitro, H1 was found to favorably undergo LLPS with nucleosomes of increasing lengths. This was unique to H1, showing that H1 could incorporate large segments of DNA within the droplets.
Another important finding was that out of the four core histones, only H2A formed droplets with DNA or nucleosomes. "The unique behavior of H2A was striking, as it meant that LLPS is not solely dependent on how much charge or structural disorder a protein has. Our results indicate that finer details of the protein structure, such as sequence and charge patterning, must be considered to predict phase behavior," says John T. King, co-corresponding author of the study. The demonstration that a core histone undergoes LLPS opens a new avenue to explore how LLPS could play a role in gene regulation and epigenetics, as histones are heavily modified by the cell to control gene expression.
The research team also explored the role of nucleotide triphosphates(NTPs) in facilitating LLPS. In particular, adenosine triphosphate (ATP), one of the cell's fuels, has been shown to contribute to the solubility of proteins and can facilitate LLPS. Indeed, the team demonstrated the essential role of ATP in promoting the formation of H1-chromatin droplets.
This work describes a new function of the histone proteins: driving LLPS with chromatin to segregate heterochromatin from euchromatin. We can imagine that many biological processes that control expression levels and biochemistry of these proteins can affect LLPS, and vice versa.
More information: Anisha Shakya et al. Liquid-Liquid Phase Separation of Histone Proteins in Cells: Role in Chromatin Organization, Biophysical Journal (2019). DOI: 10.1016/j.bpj.2019.12.022
Journal information: Biophysical Journal
Provided by Institute for Basic Science