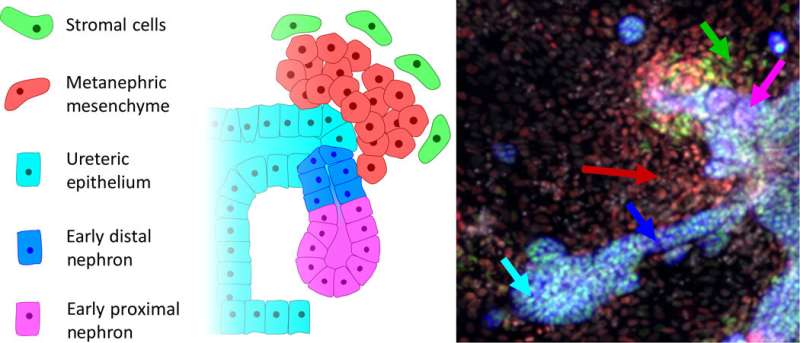
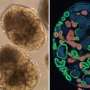
A schematic of the early developing nephron and some key cell types: Schematic of nephron development. A small budding region begins to form on the UE (blue-green). MM (red) begins to condense into clusters around the budding UE structures. These clusters of MM begin forming comma-like structures and begin to show distal (EDN, blue) and proximal polarization (EPN, magenta). This structure migrates and merges with the tubular structure of the collecting duct. Stromal cells (SM, green) also begin to appear as a step toward vascularization. Credit: Science Advances, doi: 10.1126/sciadv.aaw2746
Bioengineers have shown great promise in creating complex multicellular kidney organoids (tiny, self-organized tissues) in the lab using pluripotent stem cells . They can further improve the procedure for different outcomes, including patterning and maturation of specific cell types, although such experiments are limited by standard tissue culture approaches. Now writing in Science Advances, Nick R. Glass and an interdisciplinary research team at the Australian Institute for Bioengineering and Nanotechnology, Murdoch Children's Research Institute and the Department of Pediatrics and Biomedical Manufacturing in Australia detailed a new full factorial microbioreactor array-based method to perform complex differentiation in the lab.
Using the method, they rapidly interrogated and optimized the complex cell differentiation and development process in a simplistic setup. Glass et al. successfully recapitulated early kidney tissue patterning events and explored more than 1000 unique conditions to achieve near-pure renal cell type specification. The team conducted single-cell resolution identification of distinct renal cell types within multilayered kidney organoids and coupled the results with multivariate analysis to define the roles of several signaling proteins. The bioengineers also noted retinoic acid (RA) as a minimal effect of nephron patterning, while highlighting contributions of induced paracrine factors on cell specification and patterning.
The mammalian kidney is formed during early embryonic development from two different stem cell types; one to generate collecting ducts and the other to form functional filtering units of the kidney known as nephrons. More specifically, the mammalian kidney is a byproduct of the intermediate mesoderm (IM), arising through inductive interactions between several key progenitor (stem cell-like) IM subpopulations. While the anterior IM-derived epithelial nephric duct formed the ureter and collecting ducts of the kidney. The filtering units of the kidney or nephrons typically arose via epithelial transformations of the posterior IM known as the metanephric mesenchyme (MM). Scientists have studied normal mammalian kidney development to produce research protocols for directed differentiation of human pluripotent stem cells (hPSCs) and engineer kidney cell types to understand kidney disease. While most protocols formed primitive patterns of the trunk mesoderm, further procedures resulted in early nephron generation in the lab.
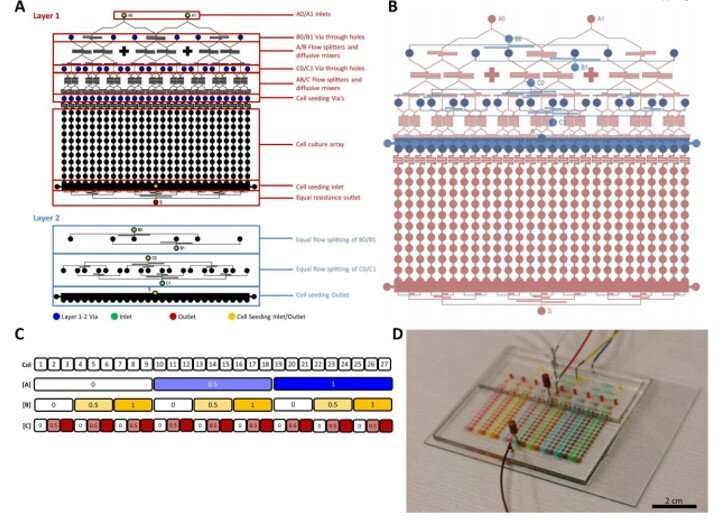
MBA design, layout, and operation. (A) The layout of the layers of the two layer MBA PDMS device showing the inlets, outlets, diffusive mixers, flow splitters, cell culture array and cell seeding ports. Colored circles are shown for holes for vias (blue), inlet (green), outlet (red) and cell seeding (yellow). These are all punched with a 0.75 biopsy punch manually. (B) The two multilayers shown in assembly format with Layer 1 (red) below layer 2 (blue). (C) A typical experimental layout of a bioreactor with the column number and working concentrations of factor A, B and C. (D) The device under perfusion with various food dyes with inlet A, B and C loaded with blue, yellow and red dyes, respectively. Photo credit: N. Glass, University of Queensland, Australia. Credit: Science Advances, doi: 10.1126/sciadv.aaw2746
For instance, after seven days of monolayer culture under three-dimensional (3-D) culture conditions, cells could self-assemble to form kidney organoids containing segmented nephrons, collecting duct epithelium, endothelial progenitors and a surrounding interstitium. Static culture conditions did not, however, allow investigators to probe direct, synergistic or paracrine influences governing cell fate specification, therefore they used expensive infrastructure such as automated liquid handling-based conditions in the lab.
Even still, the intrinsic static nature of the devices remained limiting, so to overcome limits, they used microfabricated devices with successful applications across classical biological experiments; facilitating unprecedented investigations into complex biological phenomena. Research teams had also previously designed microdevices to multiplex (amplify) experiments and simultaneously conduct many experiments on a single device. These microbioreactor arrays (MBAs) offered them opportunity to increase parameter space and reduce timeframes of protocol optimization.
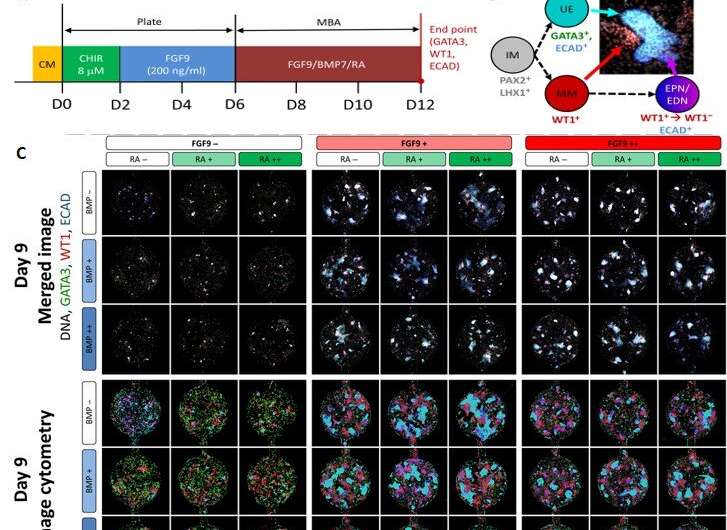
Image cytometry comparison for the first row of the day 9 and day 12 bioreactors. (A) The experimental time course setup for the day 12 bioreactors. Here, cells were cultured in well plates for the first 6 days and in the MBA system for the following 6 days and stained for DNA (white), GATA3 (green), WT1 (red), and ECAD (blue). (B) A representation of two of the kidney cell subtypes being investigated: the MM (WT1+) and the UE (GATA3+ and ECAD+) and their immunofluorescent representation. (C) The immunofluorescent imaging of the first row of a day 9 MBA array with the variation of FGF9, BMP7, and RA as indicated and the corresponding image cytometry. Wells are stained with GATA3 (green), WT1 (red), and ECAD (blue). Here, the UE (GATA3+ECAD+), MM (WT1+), EPNs (WT1+ECAD+), EDNs (ECAD+), and mesangial cells (GATA3+) can be identified, and small perturbations in the relative amounts of each phenotype can be seen. (D) The immunofluorescent imaging of the first row of a day 12 MBA array with the variation of FGF9, BMP7, and RA as indicated and the corresponding image cytometry with immunostaining as above. Credit: Science Advances, doi: 10.1126/sciadv.aaw2746
In this work, Glass et al. detailed applications of the MBA-based method to achieve unbiased, factorial assessment of stem cell-derived IM progenitor differentiation in to multiple renal cell lineages, followed by patterning them into multicellular kidney organoids. They developed a custom post-processing algorithm to quantitatively analyze the outputs of image cytometry to their phenotype—at single cell resolution. The new MBA platform validated previous protocols on renal lineage regeneration and identified previously unappreciated critical impacts of paracrine signalling during the process. Glass et al. specified roles for bone morphogenetic protein 7 (BMP7) and prolonged Wnt activation (a group of signal transduction pathways) during renal cell fate decisions and organoid patterning.
During the experiments, the team seeded the MBA with six-day old IM progenitors to produce human embryonic stem cells (HES3 cells) and treated the culture with a Wnt agonist and other nutrients. After cell culture for 16 hours, Glass et al. investigated the IM progenitors via gene expression studies and observed cell aggregates that formed a colony-like structure within the wells in time. The team then tested the influence of three molecules; fibroblast growth factor (FGF9), bone morphogenetic protein 7 (BMP7) and retinoic acid (RA) due to their previous roles in kidney cell type induction. However, the combined interplay of these factors remained unknown.
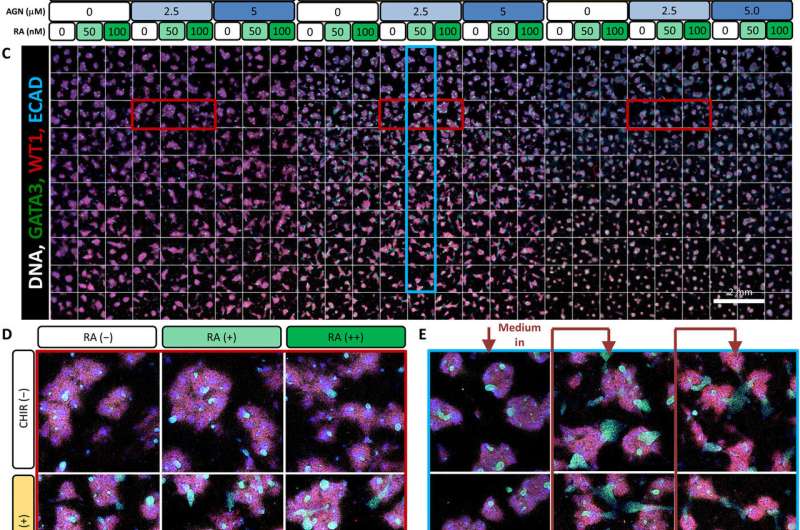
Analysis of the effect of continued Wnt signaling and RA activity after variation in the initial duration of canonical Wnt patterning (4 days of initial CHIR, day 4 to 9 MBAs). (A) Experimental plan for 4-day CHIR induction with 5-day factorial analysis (day 4 to 9 MBAs). FGF9 was present across the entire 5-day period during the factorial perfusion. (B) The working concentrations of CHIR, AGN, and RA used for both MBA experiments (day 3 to 8 and day 4 to 9). (C) The full scan of each well of day 4 to 9 MBAs stained for GATA3 (green), WT1 (red), and ECAD (blue). (D) Representative wells from row 3 representing the effect of CHIR and RA with constant AGN. These wells are highlighted about in red on the overall scan. (E) Representative wells from column 14 representing the induced paracrine signaling. These wells are highlighted in blue on the overall scan. Credit: Science Advances, doi: 10.1126/sciadv.aaw2746
Using the nested resistive network within the MBA, Glass et al. split and recombined input media of the three factors (FGF9, BMP7 and RA) at different concentrations for a total of nine or 12 days. At both timepoints, they fixed the cells, counted them and identified them (phenotyped them) using antibody labeling to readily identify distinct cell types belonging to the early mammalian embryonic kidney. The 12-day culture led to significantly more complex and mature structures. The team conducted multivariate regression analysis to understand the influence of the three factors; showing FGF9 as a vital factor for cell proliferation and maintenance of all cell types.
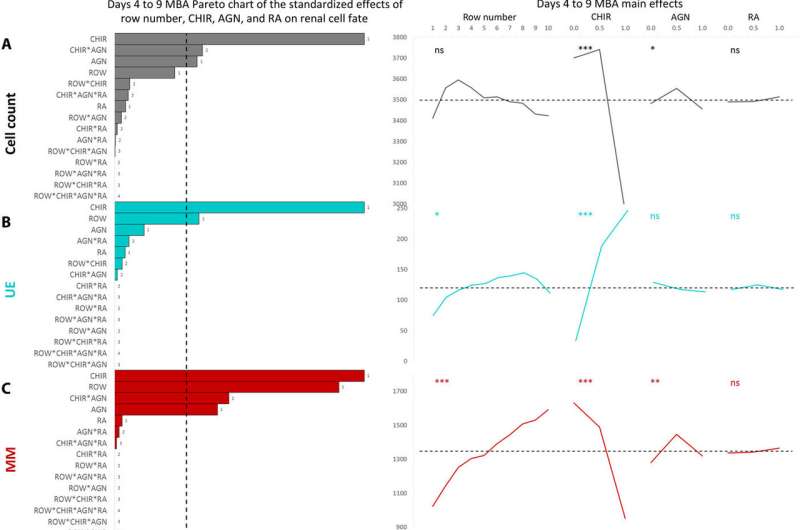
Similarly, BMP7 increased the total cell number while affecting the proportion of cell specification during early kidney differentiation. Based on the levels of BMP7 in the presence of FGF9, and in response to paracrine signaling, Glass et al. recorded substantial variations in the cell morphology (structure) within the wells. However, RA did not appear to play a role during early kidney differentiation, which apparently depended on the short, two-day exposure to WNT activation at the onset of cell culture.
Main effects plots from a full factorial design of experiments multivariate analysis for day 3 to 8 and day 4 to 9 MBAs. Pareto charts of all factors (row number, FGF9, BMP7, and RA) and their combinations and main effects of each input factor for the day 4 to 9 MBAs (n = 4). Phenotypical cell assessment included the cell count (A), UE (B), MM (C), EPN (D), EDN (E), and SM (F). The dashed line on each Pareto chart represents (P = 0.05), whereas the dashed line on the main effect plots represents the global mean of that parameter. *P < 0.5, **P < 0.01, and ***P < 0.001. Credit: Science Advances, doi: 10.1126/sciadv.aaw2746
In this way, Nick R. Glass and colleagues showed the capacity to precisely direct stem cell differentiation in the lab to form kidney tissue and individual kidney cell types as a major step towards applications in regenerative medicine and drug discovery. Since paracrine-mediated differentiation events which direct renal cell type specification can be difficult to replicate in standard static culture conditions, Glass et al. used an MBA (microbioreactor array) platform to investigate renal cell fate. The data showed that small-step changes in exogenous FGF9, BMP7 and CHIR, as well as endogenous signaling in the form of paracrine and autocrine factors, could shift the ratio of cell differentiation.
As a result, they formed ureteric, metanephric mesenchyme (MM), early proximal nephrons (EPNs) and distal nephron (DN) phenotypical cells, starting from IM precursors. Additionally, FGF9 signaling was critical to maintain all cell populations. The step-wise patterning that occurred in the MBA, was similar to the processes in the embryo, where multiple, distinct cell types that signal to each other arose across time. The MBA-based factorial approach can be applied to obtain complex, multistep differentiation for multicellular outcomes and the process will be able to tune differentiation of other organoid-like pluripotent stem-cell derived tissues.
More information: Nick R. Glass et al. Multivariate patterning of human pluripotent cells under perfusion reveals critical roles of induced paracrine factors in kidney organoid development, Science Advances (2020). DOI: 10.1126/sciadv.aaw2746
M. Takasato et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney, Nature Cell Biology (2013). DOI: 10.1038/ncb2894
Minoru Takasato et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis, Nature (2015). DOI: 10.1038/nature15695
Journal information: Science Advances , Nature Cell Biology , Nature
© 2020 Science X Network