Biophysicists find 'extra' component in molecular motor
Researchers from the Moscow Institute of Physics and Technology have discovered an additional component in ATP synthase, a molecular machine that produces the energy-conserving compound in all cellular organisms. The new unique features of the ATP synthase structure are described in detail in a paper in Scientific Reports.
In order to store energy, living cells rely on a molecule called ATP. It is produced by ATP synthase, a molecular-scale motor comprised by a rotor and a stator. Such machines are nested in the inner membranes of mitochondria and chloroplasts in most organisms, including animals, plants, and bacteria. The rotor component resembles a barrel embedded into a biological membrane. This "barrel," or C-ring, is made of between eight and 17 so-called protomers. Their exact number depends on the organism.
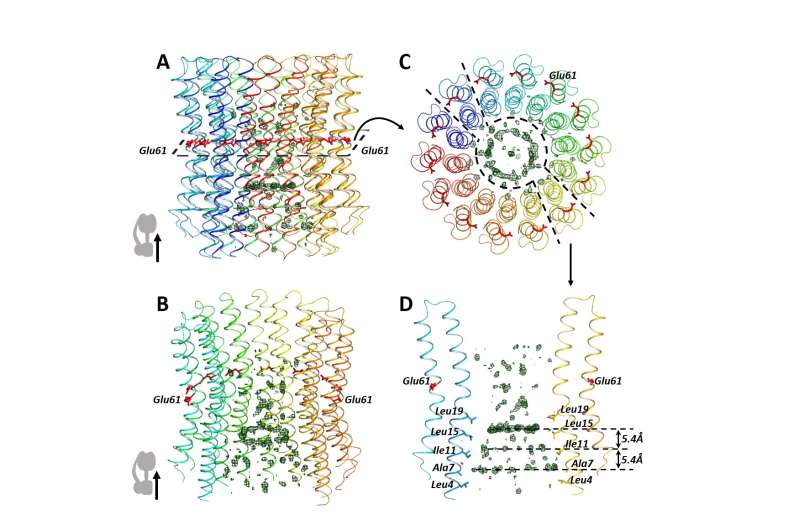
MIPT researchers and their colleagues from Grenoble, France, have obtained a first-ever high-resolution structure of the C ring from spinach chloroplasts. As the 3-D computer model of the C ring was taking shape, the biophysicists spotted something peculiar.
"We noticed additional circle-shaped elements inside the C ring," said MIPT doctoral student Alexey Vlasov from the Institute's Research Center for Molecular Mechanisms of Aging and Age-Related Diseases. "At first we thought that was an artifact. But when we looked through the C ring structures obtained by other scientists for various organisms, the circles turned up again, time after time."
It came as a surprise for the researchers that previous studies did not pay attention to the circles inside C rings. Up until now, their nature remained unexplained.
"This study speaks to the fact that no minor detail is negligible in science. Even a subtle feature, spotted in due course, might lead to a breakthrough discovery," noted Valentin Gordeliy, who heads research groups at the Institute of Structural Biology in Grenoble (France) and Jülich Research Center (Germany) and is the scientific coordinator of the MIPT Research Center for Molecular Mechanisms of Aging and Age-Related Diseases.
The biophysicists from MIPT set out to solve the C ring puzzle. Computer modeling and biochemical experiments indicated that the ring contained quinone molecules. They act as electron carriers in biological systems. Some of the examples are plastoquinone, found in chloroplasts, and the coenzyme Q in mitochondria.
Biologists have long known that the C ring of ATP synthase does not have a "hole" in it. So while some molecules were expected to exist on the inside, no one was sure which exactly. The finding proved unexpected: quinones.
While the discovery is interesting in and of itself, researchers have yet to determine why the C ring hosts quinones and how they get there. One theory suggests C rings can function as pores in mitochondrial membranes. Such a pore might open when the cell death process is initiated. Can the quinones in a C ring kill a cell? This is a question for the MIPT biologists to address in their further research.
More information: A. V. Vlasov et al, Unusual features of the c-ring of F1FO ATP synthases, Scientific Reports (2019). DOI: 10.1038/s41598-019-55092-z
Journal information: Scientific Reports
Provided by Moscow Institute of Physics and Technology