A new approach to reveal the multiple structures of RNA
Experimental data and computer simulations have yielded an innovative technique to characterize the configurations of an RNA molecule. The work, published in Nucleic Acids Research, opens new roads to studying dynamic molecular systems.
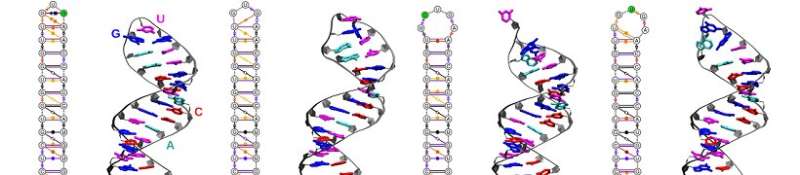
The extraordinary functionality of ribonucleic acid, better known as RNA, is due to its highly flexible and dynamic structure. Yet, the experimental characterization of its configurations is complex. A study conducted by SISSA and published on Nucleic Acids Research combines experimental data and molecular dynamics simulations to reconstruct the multiple dominant and minority structures of a single RNA fragment, providing an innovative method to studying highly dynamic molecular systems.
"RNA plays a central role in the process of protein synthesis and in its regulation. This is thanks to a structure that varies dynamically from one state to another," explains Giovanni Bussi, physicist at SISSA—Scuola Internazionale Superiore di Studi Avanzati and principal investigator of this work. "It is a set of configurations with a dominant structure maintained by the molecule for the majority of the time, and some low-population structures that are rarer, but equally important for its function and possible interaction with other molecules."
Experimental techniques, such as nuclear magnetic resonance (NMR), would theoretically allow researchers to probe these sets of configurations. However, to obtain the structure of the molecule, the assumption is often made that a single relevant conformation exists. Unless particularly sophisticated experimental approaches are used, they provide the "average" structure—in short, a mathematical average of the multiple states, which, in dynamic systems, does not exist in reality.
Giovanni Bussi and Sabine Reißer, in collaboration with the group of the neurobiologist Stefano Gustincich at the Italian Institute of Technology (IIT), have developed a new way to identify the configurations of an RNA molecule. "We studied a fragment of RNA with a characteristic hairpin structure that was identified in Gustincich's group and has an important role in regulating protein synthesis. By combining the NMR data with molecular dynamics simulations, we have mimicked different states of the molecule, including those of low population, and identified those that were able to reproduce the 'average' structure determined empirically."
The scientists then searched the Protein Data Bank, a database of molecular structures acquired experimentally: "We found some of the low population configurations identified by the simulations within other RNA molecules, including the ribosomes, the molecular machines responsible for protein synthesis ribosomal RNAs. Thus, we confirmed the existence of these multiple structures in nature and suggested a potential role at the ribosomal level," concludes Bussi. "We have demonstrated the validity of this innovative approach, which opens new roads for the study of molecular structures in highly dynamic systems in which states with different populations co-exist."
More information: Sabine Reißer et al, Conformational ensembles of an RNA hairpin using molecular dynamics and sparse NMR data, Nucleic Acids Research (2019). DOI: 10.1093/nar/gkz1184
Journal information: Nucleic Acids Research
Provided by International School of Advanced Studies (SISSA)