Realization of new image-based structure analysis method for 3-D structural analysis of biology
In basic biology, medical sciences and pharmaceutical sciences, structural analyses of membrane proteins are important, and single-particle analysis by cryoEM is now very powerful method. Japanese researchers have developed the image-based structure analysis (IBSA) method, an improved method of single-particle analysis that enables structural analysis in a lipid membrane. This development has made it possible to analyze the structures of membrane proteins in vivo.
Single-particle analysis, a cryo-electron microscopy method, does not require crystallization of the sample as in X-ray crystal structure analysis, and there is no upper limit on the molecular weight of the sample as in nuclear magnetic resonance spectroscopy. Since atomic resolution can be achieved with very few aqueous samples, single-particle analysis has established a position as one of the basic technologies for structural analysis.
However, this method requires a large number of images of randomly placed target protein particles. Purification of proteins is necessary in single-particle analysis and in the case of membrane proteins, detergent solubilization is required for this purification. This changes the molecular structure. Therefore, it is difficult to analyze the structure of membrane proteins in a real environment.
Research on the correlation between structure and function of membrane proteins requires a method that can elucidate their three-dimensional structure at the molecular level in the cell membrane. A method for resolving this problem could be the IBSA method invented by Professor Fujiyoshi of Tokyo Medical and Dental University. It captures tilted and non-tilted images of the sample, improving the reliability of 3-D reconstruction while taking advantage of single-particle analysis. It is a method of analyzing the structure of proteins in real biological membranes.
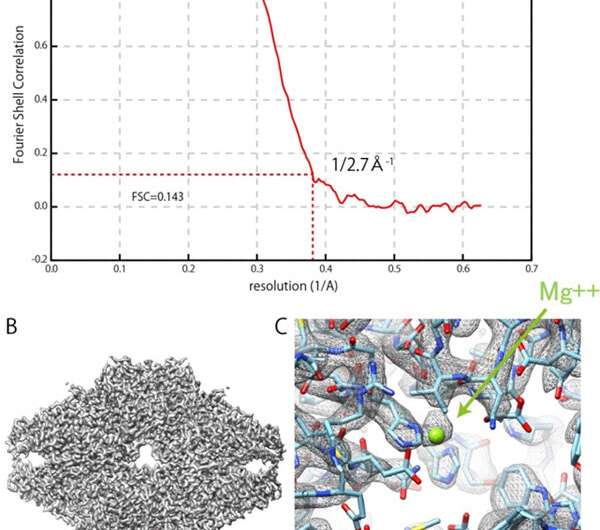
For this project, JEOL produced a cryo-electron microscope with a cryo-stage equipped with a sample autoloader and an automated liquid nitrogen supply system. For the IBSA method, the algorithm was constructed and verified, and sample preparation conditions were verified. Structural analysis using a cryo-electron microscope with the IBSA method obtained a resolution of 0.27 nm for β–galactosidase (which is an enzyme), by FSC (Fourier Shell Correlation) evaluation (FSC = 0.143).
Structural analysis with a cryo-electron microscope does not require the sample to be crystalline, unlike X-ray crystal structure analysis. Therefore, it enables structural analysis of proteins that cannot produce crystals. By using a cryo-electron microscope with the IBSA method, the structure of membrane proteins in a biological membrane can be analyzed, which could elucidate physiological functions of membrane proteins and drug discovery research.
Provided by Japan Science and Technology Agency