Growth Arrest Specific 7 protein allows cells to eat
Without hands or feet, a single human cell has little to protect itself from surrounding threats like bacteria or viruses. In this way, the cell membrane has evolved to be much more than a wall that keeps cell content inside and everything else outside. By dynamically changing its shape, the membrane allows the cell to purge itself of waste and also attack nearby threats. Japanese scientists now report how the assembly of the protein Growth Arrest Specific 7 (GAS7) into two-dimensional sheets is crucial for this second process.
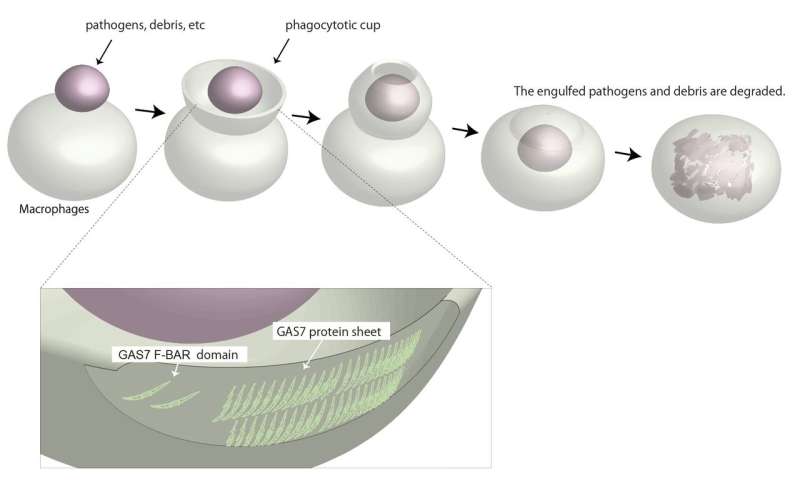
Phagocytosis describes a universal mechanism through which cells consume debris from the outside. The cell membrane will deform into a curvature known as phagocytic cup that rapidly wraps around nearby debris. Once fully engulfed, the debris will be passed into the specific organelles that will dispose of it. Immune cells such as macrophages use this process to consume and destroy infected or cancerous cells.
GAS7 is expressed in many cell types, but its function in membrane deformation is only partially understood.
"GAS7 is expressed in macrophages, and macrophages highly depend on phagocytosis, but we don't know the function of GAS7 in these cells," explains Nara Institute of Science and Technology Professor Shiro Suetsugu, who led the study.
Even though macrophages are only 20 or so microns wide, the phagocytic cup themselves measure in microns. In other words, the membrane deformation required for phagocytosis makes the cell like a snake that dislodges its jaw to eat something as small as a mouse or as big as a cow.
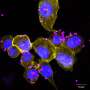
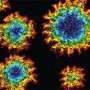
The study shows that the presence of GAS7 could mark which sites on the macrophage membrane would deform into phagocytic cups. By exposing macrophages to zymosan, a type of glucan that scientists use to model microbes, the researchers found GAS7 along with an assortment of other molecules indicative of the beginnings of phagocytosis accumulated.
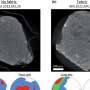
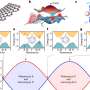
Of particular interest to Suetsugu was a specific region in GAS7, the Bin-Amphiphysin-Rvs167, or BAR, domain. BAR domains come in several types and are responsible for a protein's ability to bend the cell membrane. Crystal structures revealed a distinctive morphology of the GAS7 BAR domain to be the F-BAR domain with small protrusion on its surface.
"These domains formed filamentous oligomers with flat surfaces. The oligomers had an asymmetry due to two GAS7-specific loops so that one side is more positively charged than the other. The positively charged side, we supposed, could be the membrane-binding surface," says Suetsugu.
Mutants in these loops compromised the macrophage phagocytosis for zymosan.
"Our findings indicate that the oligomerization and membrane binding of GAS7 are essential for phagocytic cup formation," continued Suetsugu.
GAS7 is just one of many but essential proteins that have evolved to support phagocytosis. Elucidating the roles of the many proteins involved in this universal mechanism will explain not only the dynamics of the membrane, but also insights on how cells could thrive and thus grow into multicellular organisms.
"Further analyses will clarify how GAS7 oligomers cooperate with other proteins to regulate membrane deformation in phagocytosis," says Suetsugu.
More information: Kyoko Hanawa-Suetsugu et al. Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7, Nature Communications (2019). DOI: 10.1038/s41467-019-12738-w
Journal information: Nature Communications
Provided by Nara Institute of Science and Technology