New structure for human flu virus protein

Researchers from Oxford University have worked out the molecular structure of a protein that is vital for survival of the flu virus. Recently published in Nature, they used several different techniques to look at the arrangement of atoms within a protein that the human flu virus uses to make new copies of its genetic information. Without this multifunctional protein, known as a polymerase, the flu virus cannot survive. A key finding of the study is that the polymerase can exist in two forms, a monomer and a dimer. It is only when the polymerase dimerises that specific functions are switched on. The research team saw the dimeric form of the polymerase was formed by a specific region of protein and when the region was disrupted, the polymerase couldn't work. This finding presents a brand-new way of potentially inhibiting the flu virus which means we could develop new drugs and flu treatments in the future.
A structure and function for an important influenza protein
Structural biology has been used to understand important human diseases for decades. By understanding cellular interactions on a molecular level, it is possible to find treatments and cures. However, we lack a lot of structural information for many functionally important proteins. Influenza A virus is an avian virus, which is able to jump species, infecting both humans and other animals. Influenza A viruses are the most common cause of seasonal flu in humans and cause annual epidemics around the world and occasional pandemics, representing significant public health and economic burdens. When influenza virus infects a host cell, it starts to make many copies of itself as the disease spreads. The protein that drives this copying is the viral RNA polymerase which replicates the viral RNA genome and makes RNA templates for protein synthesis. In order to understand how the polymerase works in mechanistic detail you need to work out the structure of the RNA polymerase in atomic detail.
The reason that no structures existed for this protein until recently is that it was very difficult to produce in a highly purified form. In order to do structural studies, proteins normally have to be cloned and produced in large quantities in bacteria, and sometimes it's just not possible. However, several years ago, the groups of Ervin Fodor and Jonathan Grimes, along with others around the world, discovered a way to produce the flu polymerase in large amounts. Then the race was on to determine the structure.
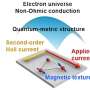
Fast forward to today and the team has published the structure in the journal Nature. In the article they describe a high-resolution structure of the RNA polymerase in multiple conformations which shows how the protein is able to replicate the influenza virus RNA genome. They observed that the protein could either form monomers or dimers. Grimes explained, "We saw that the polymerase existed as a mixture in two forms, as monomer and a dimer. This is quite unusual, but it was not clear whether this observation had any functional implications."
Ervin Fodor had previously published work from his lab that showed that a key step in the viral genome replication required some form of activation before it could work. The team hypothesised that this trans-activation comes from formation of the dimer.
The crystal structures of the dimerised polymerase clearly showed the region of the protein that formed the interface holding the dimer together. The research team wanted to know if this region was important to maintaining the dimer and more importantly, if they could disrupt the activity of the polymerase by forcing it into a monomeric form. They mutated various parts of the dimer interface to produce a selection of mutant proteins that could not form dimers. These experiments showed that a key step in their replication of the viral genome was blocked if dimerization was prevented from occurring.
This finding was confirmed independently using nanobodies which are much smaller versions of antibodies that can still recognise specific antigens. Their use was pioneered in structural biology where they are routinely used to help with crystallisation of proteins or to provide support to "floppy" structures during cryo-electron microscopy (Cryo-EM). However, their small size also makes them perfect for molecular biology studies where larger antibodies would get in the way. Fodor and Grimes collaborated with a group in Belgium who produced a number of different nanobodies that bound to the influenza virus polymerase. They found that when one of these nanobodies were added to the polymerase, it inhibited RNA replication but not the other functions involved in protein production. Solving the structure of the RNA polymerase in complex with this nanobody revealed that the nanobody bound at the dimer interface. This not only gave support to the idea that the dimer was essential for the function, but suggested that this dimer interface is a novel target for antiviral drugs.
Using a battery of different techniques from Diamond
The research was made possible by an array of different structural and molecular biology techniques, with the structural component being carried out at Diamond Light Source. The team used three different techniques, Cryo-EM, crystallography and SAXS which are powerful when used on their own, but even more so in combination when addressing complex, structural biology questions.
Cryo-EM was used extensively in this study to investigate the structure of the viral polymerase. The technique itself has been around for decades but it wasn't very useful for structural biology until recently with the introduction of new direct electron detectors. These new detectors allow the capture of very high-resolution molecular movies that allow experimenters to image their protein samples at high resolution. These detectors have led to the so called cryo-EM resolution revolution. One of the huge benefits of Cryo-EM is that you don't need to grow crystals which is a significant limitation if relying on X-ray crystallography. "Cryo-EM has allowed us to begin to look at very interesting protein complexes that we would find impossible to grow crystals of in the lab," explained Grimes.
That being said, X-ray crystallography was still an important component of the study. "X-ray crystallography and Cryo-EM generate data in completely different ways, so if possible, it's good to have both. Crystals might tell you one things and Cryo-EM another, by combining them you can complement your results," explained Grimes. Crystallography also tends to generate higher resolution data sets and until now, is faster to analyze.
The third technique to be used at Diamond was small angle X-ray scattering (SAXS). While it can be considered low resolution, especially compared to Cryo-EM and crystallography, SAXS is often used as a complement to high resolution structural techniques because it gives a dynamic view of the protein. In this case, the research teams were able to see the dimers forming in real time and be sure that they were necessary for the polymerase to function. Grimes said, "The fact that all of these techniques exist in one place and are available to the scientific community is a hugely valuable resource, Diamond democratizes science, since these world class cutting-edge facilities are freely available to scientists from universities and institutes across the UK and EU, with interesting and important biological questions."
What's next?
This is not the first time that this group has published an important protein structure. In 2015 they published the structure of the polymerase from the influenza C virus so the future could very well involve producing more important structures. However, there are also some new techniques being developed that the researchers are eager to apply. One of these is cellular X-ray and Cryo-EM tomography which means taking living cells, freezing them and then analyzing the structures of cellular structures and proteins in situ. The groups of Grimes and Fodor want to do this with the flu polymerase replicating inside a cell. While there are still significant limitations to this technique, the proof of concept exists and this would begin to address significant questions about what is happening in vivo.
The study also has immediate applications for medicine and drug discovery. The research team demonstrated a new way to inhibit the human influenza A polymerase which is a perfect target for new antiviral drugs. This discovery was possible due the exhaustive study using many different techniques.
More information: Haitian Fan et al. Structures of influenza A virus RNA polymerase offer insight into viral genome replication, Nature (2019). DOI: 10.1038/s41586-019-1530-7
Journal information: Nature
Provided by Diamond Light Source